Desulfurococcus mobilis
A Microbial Biorealm page on the genus Desulfurococcus mobilis
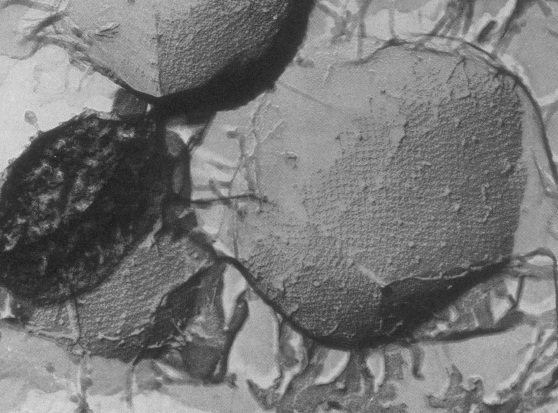
Classification
Domain: Archea; Phylum: Crenarchaeota; Class: Thermoprotei; Order: Desulfurococcales; Family: Desulfurococcaceae ; Genus: Desulfurococcus; Species: mobilis.
Description and significance
Desulfurococcus mobilis is an extreme thermophile, living up to temperatures of 97° C and at a pH between 2.2 and 6.5. The species ranges in size from about 0.5 microns to 10 microns, and is covered with a unique, tetragonally-arrayed surface protein which forms a mesh of cross-shaped units. It is an anaerobe and is dependent upon sulfur for respiration. D. mobilis is found in solfataric (volcanic and sulfur emitting) hot springs and is most commonly isolated under these conditions in the country of Iceland. Its ability to survive in extreme conditions makes this archaeon valuable for uses in biotechnology, since thermostable and thermoactive enzymes can be isolated from this organism, like the restriction enzyme I-Dmol. Interestingly, the first known prokaryotic rRNA intron was discovered in D. mobilis, helping to give insights on the evolutionary relationship between archaea, bacteria and eukaryotes. Further, this organism is not a known pathogen.
Genome structure
D. mobilis contains a single and circular chromosome. It is not known to contain any plasmids. The genome of D. mobilis has not yet been sequenced, however its closest relative, Desulfurococcus mucosus was determined to have a genome size of 1.3 million bp. There exists a 622bp intron within the 23S rRNA gene with a splicing site in domain IV.
Cell structure, metabolism & life cycle
This organism is an organotroph and has a strictly anaerobic metabolism. It acquires energy by fermenting peptides. Its growth is dependent upon the reduction of sulfur and inhibited in the presence of hydrogen gas. They can be identified by their coccus shape and unique protein coat. The exterior of the organism is coated with irregularly arranged surface glycoproteins. These proteins form a lattice which is comprised of cross-shaped units. The protein coat is thought to function as a molecular sieve that mediates the adsorption of small to medium-sized molecules (>700kd). Additionally the protein coat is flexible, and possibly works as an exoskeleton to assist against osmotic stress.
Ecology
D. mobilis is found in solfataric hot springs. They are found in fumaroles, or openings within the earth's crust, which emit sulfur gas and water vapor. The temperature in these fumaroles can reach up to 97° C. It has been isolated in parts of Iceland and within the United States. Its contribution to the environment is not well-documented, however it is not thought to be pathogenic towards any organism. Interestingly, the archaeon is a host to the Rudivirus. This virus is characterized as rod-shaped, with a double-stranded DNA genome. It most often found occupying the same environment as D. mobilis, however the virus-host interaction is not well known. The Rudivirus follows a lytic cycle of virus replication and degrades the host chromosome by assembling within the cytoplasm.
Interesting feature
Although there is little known about this organism, scientists have greatly exploited the enzyme I-Dmol from Desulfurococcus mobilis for uses in biotechnology. I-Dmol is a meganuclease, which is a naturally occurring restriction enzyme that is capable of cleaving phosphodiester-bonds of a recognition site on either RNA or DNA. The recognition site for I-Dmol is extremely specific at 25bp long, which is typically only found once in any given genome. The enzyme is of use in biotechnology because it acts as a "molecular scissor" which can eliminate or modify a genomic sequence in a number of different ways. This is of particular importance in genome engineering and gene therapy. In 2008, scientists discovered a way to alter the active site of this enzyme to identify and cleave different recognition sites. The implications of the re-engineered I-Dmol could be used in gene therapy, as a way of cleaving unwanted viral DNA in a host's system.
References
[http://books.google.com/books?id=3WvYMKmIvY8C&pg=PA102&lpg=PA102&dq=^+Zillig,+W.,+Prangishvili,+D.,+Schleper,+C.,+Elferink,+M.,+Holz,+I.,+Albers,+S.,+Janekovic,+D.+and+G%C3%B6tz,+D.+%281996%29.+Viruses,+plasmids+and+other+genetic+elements+of+thermophilic+and+hyperthermophilic+Archaea.+FEMS+Microbiol.+Rev.,+18,+225-236&source=bl&ots=mYdSF1wTPp&sig=46A4ivENbv6vG4MraeKDinK-pfM&hl=en&ei=rfWkTqepD6Lh0QHLvficBQ&sa=X&oi=book_result&ct=result&resnum=1&ved=0CBsQ6AEwAA#v=onepage&q=^%20Zillig%2C%20W.%2C%20Prangishvili%2C%20D.%2C%20Schleper%2C%20C.%2C%20Elferink%2C%20M.%2C%20Holz%2C%20I.%2C%20Albers%2C%20S.%2C%20Janekovic%2C%20D.%20and%20G%C3%B6tz%2C%20D.%20%281996%29.%20Viruses%2C%20plasmids%20and%20other%20genetic%20elements%20of%20thermophilic%20and%20hyperthermophilic%20Archaea.%20FEMS%20Microbiol.%20Rev.%2C%2018%2C%20225-236&f=false Zillig, W., Prangishvili, D., Schleper, C., Elferink, M., Holz, I., Albers, S.,
Janekovic, D. and Götz, D. "Virus taxonomy: classification and nomenclature of viruses : eighth report" International Union of Microbial Sciences. 1998.]
