Diego's microbe wiki
- 'Enterobacter Cloacae'
Classification
'Kingdom': Bacteria
Phylum': Proteobacteria
'Class': Gamaproteobacteria
Order': Eubacteriales
Family': Enterobacteriaceae
'Genus':Enterobacter;
Habitat Information
Enterobacter Cloacae are typically found in nature and are capable of obtaining nutrition from products of organic breakdown and decay. They are often found in environments of stagnant water, soil, sewage, and are also located within the "normal" stomach flora of both animal and human GI tracts. (+/- 40%)
https://www.google.com/maps/@30.3133784,-97.7208394,13z
Description and Significance
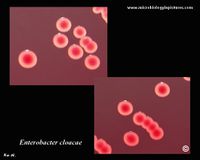
To the human eye individual colonies of the microbe appear white/creamy, with an umbonate elevation similar to an egg sunny side up. It's margins are round, smooth/entire.
Diversity of its attachment pili allows for its resilient ability to colonization on various hosts. Genetic analysis further shows that E. cloacae strains possess multiple mechanisms for antagonistic action against other microorganisms, which include the production of various antimicrobial compounds and antibiotic resistance proteins. These give the microbe further fitness advantages in microbial competition, thus allowing it to survive in different environments.
A recent study has shown that the presence of Enterobacter cloacae in the gut may have a strong correlation to obesity likelihood. A decrease 35% to non-detectable levels of the bacterial within the patient’s gut, was linked to a strong reduction in endotoxin load which significantly reduced the patient's weight. A 2012 study where Enterobacter cloacae was transplanted into previously germ-free mice resulted in increased obesity when compared with germ-free mice fed an identical diet, suggesting a link between obesity and the presence of Enterobacter gut flora.
Species
'Species':Cloacae
the suffix "Enteric" signifies a microorganism pertaining to or originating from the intestines.
Several Species of the Enterobacter cloacae complex exist, but only few act as pathogens. The biochemical and molecular studies on E. cloacae have shown great genomic similarity with six seperate species. Often referred to as Enterobacter Subspecies or 'Enterobacter spp.'
- 1) Enterobacter cloacae
- 2)Enterobacter asburiae
- 3)Enterobacter hormaechei
- 4)Enterobacter kobei
- 5)Enterobacter ludwigii
- 6)Enterobacter nimipressuralis.
E. cloacae and E. hormaechei are the most frequently found in human clinical specimens. Molecular identification is often used to differentiate between these similarly related species.
' BASIC TESTS
- ' FOR IDENTIFICATION
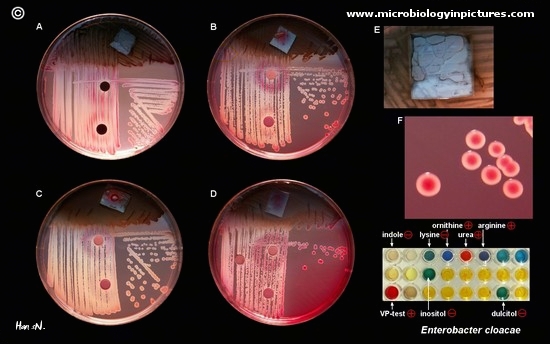
- MacConkey growth = + for growth of gram - and + for lactose fermentation
- Indole production = - for the presence of triptophan 1
- Methyl red = - for mixed acid fermentation
- Voges-Proskauer = + for neutral end products, (my lab test results came back -)
- Citrate(Simmons) = + for citrate carbon source
- Lysine decarboxylase = - for fermentation ( my lab results came back +)
- Arginine dihydrolase = + for removal of carboxyl group from amino acid
- Ornithine decarboxylase = + for decarboxylase
- Motility (36 °C) = + growth in SIM broth
- Mannitol salt test = + acid and gas production < br />
- Sucrose fermentation = + acid and gas production
- Lactose fermentation = + acid and gas production
- Bile Esculin =+ hydrolysis of esculine in presence of bile
- Antibiotic Test = + resistant to cefemandole
- MacConkey growth = + for growth of gram - and + for lactose fermentation
- ' FOR IDENTIFICATION
Cell Structure, Metabolism and Life Cycle
Enterobacter Cloacae cells are a genus of straight gram-negative bacilli (rods) which contain a thin cell-wall of peptidoglycan. E. Cloacae are capable of Nitrate reduction by removing oxygen from nitrate (NO3) producing nitrite (NO2), lactose-fermentation, are apart of the family Enterobacteriaceae and are facultatively anaerobic (rods), which have the potential to grow and multiply with or without oxygen. The average cellular size ranges from 0.6-1 μm in diameter and 1.2-3 μm long. E. Cloacae are capable of movement by means of peritrichous flagella, using multiple tails for propulsion, and are also acid producers upon glucose fermentation, with an optimal growth temperature of 30 °C. Close to 80 % of bacterial cells are encapsulated.
- GRAM-NEGATIVE RODS =pink after gram staining
- MOTILE
- NONSPOREFORMING
- CATALASE: POSITIVE
- OXIDASE: NEGATIVE
- FACULTATIVELY ANAEROBIC
Genome Structure
Close examination of the E. cloacae genome shows that it's primary genetic sequence reveals the general physiological and survival genes of the species. Genomic factors in plasmids determine the virulence of the human pathogenic E. cloacae strain.
E. cloacae chromosomal DNA consists of 40-60% guanine + cytosine base pairs (G+C) nucleotides. The complete E. cloacae subsp. cloacae genome contains a single circular chromosome of 5,314,588 bp and two circular plasmids.
- PCR sequencing forward: ATGCAGCCATGCCGCGTGNATGAAGAAGGCCTTCG
GGTTGTAAAGTACTTTCAGCGGGGAGGAAGGTGTTGTGGTTAATAACCGCAGCAATTGACGTTACCCGCAGAAGAAGCAC CGGCTAACTCCGTGCCAGCAGCCGCGGTAATACGGAGGGTGCAAGCGTTAATCGGAATTACTGGGCGTAAAGCGCACGCA GGCGGTCTGTCAAGTCGGATGTGAAATCCCCGGGCTCAACCTGGGAACTGCATTCGAAACTGGCAGGCTAGAGTCTTGTA GAGGGGGGTAGAATTCCAGGTGTAGCGGTGAAATGCGTAGAGATCTGGAGGAATACCGGTGGCGAAGGCGGCCCCCTGGA CAAAGACTGACGCTCAGGTGCGAAAGCGTGGGGAGCAAACAGGATTAGATACCCTGGTAGTCCACGCCGTAAACGATGTC GATTTGGAGGTTGTGCCCTTGAGGCGTGGCTTCCGGAGCTAACGCGTTAAATCGACCGCCTGGGGAGTACGGCCGCAAGG TTAAAACTCAAATGAATTGACGGGGGCCCGCACAAGCGGTGGAGCATGTGGTTTAATTCGATGCAACGCGAAGAACCTTA CCTGGTCTTGACATCCACAGAACTTTCCAGAGATGNNTTGGTGCCTTCNGGAACTGTGAGACAGGTGCTGCATGGCTGTC GTCAGCTCGTGCCGTGAGATGTCAT
- PCR sequencing reverse: ATGCAGCACCTGTCTCACAGTTCCCGAAGGCACCAATCCATCTCTGCGNAAGTTCTGTGGATGTCAAGA
CCAGGTAAGGTTCTTCGCGTTGCATCGAATTAAACCACATGCTCCACCGCTTGTGCGGGCCCCCGTCAATTCATTTGAGT TTTAACCTTGCGGCCGTACTCCCCAGGCGGTCGATTTAACGCGTTAGCTCCGGAAGCCACGCCTCAAGGGCACAACCTCC AAATCGACATCGTTTACGGCGTGGACTACCAGGGTATCTAATCCTGTTTGCTCCCCACGCTTTCGCACCTGAGCGTCAGT CTTTGTCCAGGGGGCCGCCTTCGCCACCGGTATTCCTCCAGATCTCTACGCATTTCACCGCTACACCTGGAATTCTACCC CCCTCTACAAGACTCTAGCCTGCCAGTTTCGAATGCAGTTCCCAGGTTGAGCCCGGGGATTTCACATCCGACTTGACAGA CCGCCTGCGTGCGCTTTACGCCCAGTAATTCCGATTAACGCTTGCACCCTCCGTATTACCGCGGCTGCTGGCACGGAGTT AGCCGGTGCTTCTTCTGCGGGTAACGTCAATTGCTGCGGTTATTAACCACAACACCTTCCTCCCCGCTGAAAGTACTTTA CAACCCGAAGGCCTTCTTCATACACGCGGCATGGCTGCATCAGGCTTGCGCCCATTGTGCAATATTCCCCACTGCNGCCN CCCGTANGAGTACTGGCG
Physiology and Pathogenesis
E. cloacae is commonly known as an opportunistic human pathogen found in hospitals and causing a wide range of infections, such as lower respiratory tract infections, urinary tract infections, and meningitis. E. cloacae often contaminates medical equipment such as hospital IV devices. Outbreaks have also been linked to colonization of certain surgical equipment and operative cleaning solutions. Outbreaks typically occur in Intensive Care Units, usually affecting patients with weakened immune systems who have been hospitalized for a prolonged periods of time. E. cloacae is significant in the clinic, mainly because its strains usually carry multiple antibiotic resistance genes. Over the last 15 years, numerous reports have revealed their remarkable ability to adapt or acquire resistance determinants and making them some of the most dangerous microorganisms of the current antibiotic era.
In plants however, Intestinal E. cloacae strains have been shown to colonize and benefit growth in various crops, such as soybean, cucumber, corn, rice and ginger.
References
Dorothy M. Hinton & Charles W. Bacon, 10 January 1995, Enterobacter cloacae is an endophytic symbiont of corn, Mycopathologia 129: 117-125, 1995. 117 © 1995 Kluwer Academic Publishers. Printed in the Netherlands. USDA!ARS, Toxicology and Mycotoxin Research Unit, Russell Research Center, Athens, Georgia, USA
http://www.microbiologyinpictures.com/enterobacter%20cloacae.html
http://www.slideshare.net/AliaNajiha1/enterobacteriaceae-basic-properties
http://www.medscape.com/viewarticle/768204
Author
Page authored by Diego M. Escobedo, student of Prof. Kristine Hollingsworth at Austin Community College.