Domestication of Canis lupus familiaris and Subsequent Transformation of Gut Microbiome
Introduction

Dogs (Canis lupus familiaris) descended from wolves (Canis lupus) thousands of years ago through domestic interactions with humans. Understanding the divergence of dogs from wolves proves challenging as analytical results provide confounding evidence to the demographic and temporal origin of domestication. The domestication of dogs by humans is associated with a genetic bottleneck resulting in a reduction of genetic diversity when compared to their wolf ancestors. Incomplete lineage sorting creates phylogenetic gaps in wolf and dog lineages. Wolves and dogs are capable of interbreeding, causing regionally restricted admixture during domestication and post-divergence gene flow between dogs and wolves. Finally, the divergence of dogs is relatively recent in evolutionary terms which explains why polymorphisms may still be shared between dogs and wolves making morphological analysis challenging.[2]
The domestication process likely occurred in three stages. The earliest dogs most likely associated with hunter-gatherer communities as opposed to agrarian cultures. These early dogs scavenged carcasses and scraps left behind by humans. In this stage of domestication, selection may have still favored a wolflike morphology, producing wolflike dogs. As humans settled and became sedentary, selection for smaller and phenotypically diverse canids became more favorable, producing more doglike phenotypes. The last stage of domestication occurred in the Victorian era where artificial selection for novelty traits created a vast range of phenotypes.[2]
Geographic and Temporal Origins of Domestication
The goal of archaeological, morphological, and phylogenetic analysis of dogs and wolves is to establish a demographic and temporal origin for the domestication of dogs. Understanding where and when dogs evolved provides key insights into both the history of contemporary dogs, as well as human history and its interaction with these animals.
Fossil Record
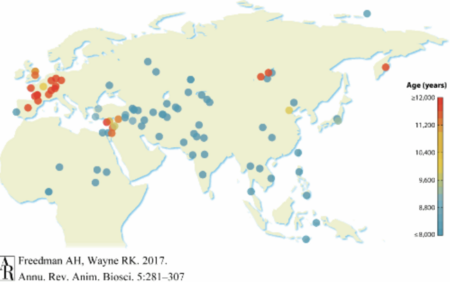
Archaeological analysis of the fossil record is challenging because early dogs are morphologically similar to wolves, making them difficult to distinguish and accurately document. Additionally, wolves were geographically broadly distributed making the origin of domestication difficult to determine. Furthermore, canid fossils are rare, causing fragmentation in the fossil record. Despite these challenges, fossil remains of wolves associated with humans can be identified as early as 400 thousand-years-ago (kya). The appearance of dogs is much more recent with one of the oldest fossils found in the Altai Mountains in Russia dating to approximately 33 kya. Many fossils have been reclassified over time as wolves or as dogs, but overall fossils of both canids can be found across Eurasia. The fossil record demonstrates a complex evolutionary process that may contain morphologically wolflike dogs and doglike wolves.[2][3]
Phylogeny
Mitochondrial DNA and Y Chromosome Sequencing
The initial domestication process of dogs is associated with a genetic bottleneck that reduced genetic variation in comparison to wolves. It caused a dramatic imprint on the dog genome resulting in long regions of auto zygosity. To identify the temporal and geographic origin of dog domestication, researchers focused on dog genomes from different geographic locations. They identified genomes with greater diversity which possibly suggests an older point of origin. This is because, over time, genetic variation in dogs increased through mutations and breeding. The more diverse the dog genome is, the more time it likely took to develop. The following includes multiple independent studies where researchers used different genetic tools to identify where and when dogs were originally domesticated.[2]
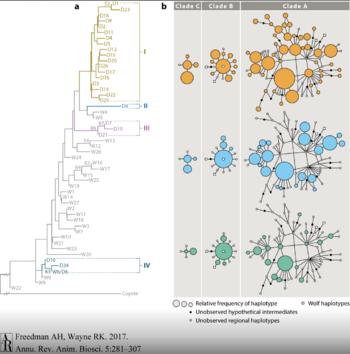
Early studies analyzed single locus mitochondrial DNA (mtDNA) and Y chromosome diversity in modern-day dogs and wolves. This reduced genetic recombination in their study to better identify the origin of domestication. This is because mtDNA is only inherited from the mother and the Y chromosome is only inherited from the father. This creates a clear record of the maternally and paternally lineages. By comparing control region sequence data between dogs and wolves, four major clades containing dog haplotypes were identified (Fig. 3). Dogs in Clade A contained the majority of unique dog haplotypes while dogs in the other three clades shared both dog and wolf haplotypes. The shared haplotypes were mostly identified in wolves from Europe. The presence of four clades may indicate multiple origins of domestication, or a single origin residing in clade A and the migration with repeating rounds of hybridization.[2][5]
Another study analyzed the clades in more detail and discovered wolf haplotypes in clade A originating in China and Mongolia. This may suggest that dogs came from East Asia. Further study of modern dog mtDNA sequences from this region revealed a higher mean sequence divergence compared to dogs from Southwest Asia and Europe. The greater genetic variation in East Asia may indicate an older point of origin which may date back to the domestication of the dog.[2][6]
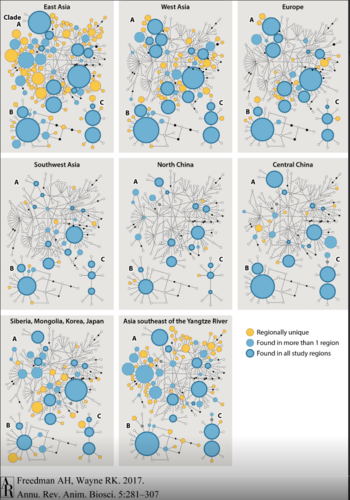
More research, sampling modern dogs across the Old World, found that 97.4% of the dogs had haplotypes from clades A, B, or C. In the sampled dogs, there was a large proportion of universal control region haplotypes which would propose a common-source gene pool. However, the majority of regional specific haplotypes were found in dogs from Asia Southeast of the Yangtze River (ASY). Additionally, every haplotype from dogs studied outside of this region could be traced to one from ASY, but the reverse was not true (Fig. 4). This study provided more evidence of increased genetic diversity in East Asia, suggesting a point of origin for dog domestication.[2][7]
In another approach to identify the origin of dogs, researchers compared the genetic diversity of mtDNA hypervariable regions (D-loop), as well as single tandem repeats and single nucleotide polymorphisms (SNPs) from the nonrecombining region of the Y chromosome between dogs from Southeast Asia and the Middle East. The mtDNA D-loop sequences reflected similar diversity in Southeast Asia as earlier studies of mtDNA control regions. When comparing haplotypes from the Y chromosome, dogs from the Middle East had more universal haplotypes while dogs from Southeast Asia had haplotypes that were unique and not found in other regions. Overall, this study concluded similar findings that dogs in Southeast Asia were more genetically diverse, supporting an older point of origin. However, the authors also argued that their findings suggest that Middle Eastern dog ancestry was influenced by immigrant lineages from Southeast Asia.[2][8]
Other research expanded their scope of study from Eurasia and sampled African village dogs from across the African continent. They analyzed mtDNA D-loop sequences and single nucleotide polymorphisms. African village dogs showed considerable genetic diversity with contributions from ancient Middle Eastern and Southeast Asian dog lineages. While Southeastern Asia remains a likely primary center of domestication, the data from African village dogs suggests a more complex model of the origin of dog domestication. This model explains the importance of post-domestication dog migrations and regional admixture in shaping modern dog populations. This study underscored the need for broader sampling populations for a more accurate representation of dog origins. This is because dog domestication was likely not a single linear event but a culmination of multiple points of origin.[2][9]
Then, genomic sampling was extended further analyzing single nucleotide polymorphisms in supposed ancient dog breeds (whose lineages extend beyond contemporary dog breeds) and wolves from the Middle East. This study used a slightly different approach, identifying that Middle Eastern wolves demonstrated a larger fraction of unique dog haplotypes than any other wolf population, including that of a Chinese wolf. The authors argued that Middle Eastern wolves contributed more genetic diversity to modern day dogs and that contributions from other wolf populations was more breed specific.[2][10]
A cumulative study analyzed the genetic variation in the complete mtDNA genome in prehistoric canids from Eurasia and the New world, as well as a panel of modern dogs and wolves. Clades A, C, and D shared 78% of dog mtDNA with the genomes sampled. The most diverse of these clades was claded A. The closest sister group for dogs in each clade were traced back to European canid origins. The association of sequences with European wolves argues for an origin of domestication in Europe. [2][11]
Whole Genome Sequencing
With multiple demographic locations hypothesizing the origin of dog domestication, future studies on the time period that dogs were domesticated needed to avoid a geographic sampling bias. They sampled dog breeds and wolves from the Middle East/Africa, southeast Asia, and Europe. Through whole genomic analysis, they concluded that none of the extant wolves from the three hypothesized regions was the source of dog domestication. Their findings suggested that dogs descended from a now-extinct wolf populations approximately 11-34 kya.[2]
A final study used regionally specific post-divergence gene flow as a model to estimate the mutation rate of dogs to validate the temporal origins of domestication. They compared the probability of a derived allele in a 35 kya Taimyr wolf sample to a heterozygote in a modern sample. They used the age to calibrate the mutation rate and concluded that previous temporal estimates should be shifted later to approximately 27-40 kya.[2][12]
Ultimately, the origins of dog domestication remain hypothetical. However, there is many sources of evidence that the origins of dog domestication are located in Europe, the Middle East/Africa, and Southeast Asia. Additionally, it is estimated that dogs diverged from wolves approximately 11-40 kya.[2]
Contemporary Morphometry
Long after the initial domestication process of dogs, largely in the recent 200 years, there has been rapid phenotypic radiation through artificial selection and breeding. This has produced immense genetic variation ranging from the one-pound poodle to the two-hundred-pound mastiff. The size, body, limbs, and skull proportions vary greatly among breeds. A great example of this is the elongated body of the dachshund. Contemporary society continues to select for additional characteristics, increasing genetic diversity, like hair length, tail curling, and coat curling.[2]
Gut Microbiome
Dogs fulfilled a new biological niche after being domesticated from wolves. The transformation of their gut microbiome is reflective of their dietary shift and adaptation to human environments. The closest extant relative of the dog (Canis lupus familiaris) is the gray wolf (Canis lupus), making the gray wolf a primary subject of comparative studies. Many organisms rely on microbes in the gut to properly digest food. When comparing captive wolves fed uncooked meat and dogs fed commercial dog food, there was a striking difference in abundance of bacteria communities in the gut.[13]
Canis lupus
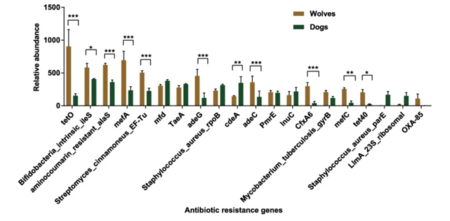
The gut microbiota of wolves was enriched with genes involved in lipid and protein metabolism, as well as genes conferring resistance to antibiotics used in livestock. Clostridium was the most abundant bacteria identified in the wolf gut microbiome which facilitates lipid and protein metabolism (Fig. 6). Shotgun metagenomic sequencing revealed genes conferring resistance to tetracycline, mupirocin, novobiocin, macrolide, elfamycin, and cephamycin (Fig. 5). Farms often use antibiotics to facilitate the growth of crops and livestock. Antibiotics are also used to treat meat products harvested from livestock. These antibiotics can be indirectly transferred to wolves who consume uncooked meat.[13]
Canis lupus familiaris
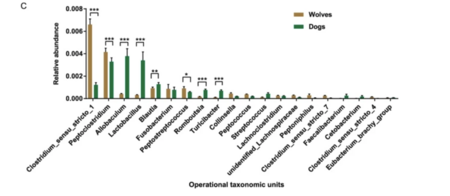
Dogs demonstrated a more diverse community of gut bacteria when compared to the wolf which had genes providing resistance to clinical antibiotics (Fig. 6). Genes conferring resistance to cephalosporin, methicillin, and sulfamethoxazole were identified in the metagenomics of the gut microbiota (Fig. 5). This resistance to clinical antibiotics may come from probiotics incorporated in commercial dog food to better dog health. Dogs also receive domestic veterinarian treatment and direct antibiotics to maintain health, impacting the communities of bacteria in the gut.[13]
The gut microbiota of dogs was enriched with genes involved in lipid and protein metabolism, as well as starch and carbohydrate fermentation. Allobaculum, which facilitates lipid and protein metabolism, and Lactobacillus, which facilitates carbohydrate fermentation, was most abundant in the dog gut microbiome (Fig. 6). A copy expansion in the gene AMY2B was observed, indicating enhanced starch metabolism. Glycosyl hydrolase enzymes break glycosidic bonds between complex carbohydrates. Genes encoding these proteins were in higher abundance in the microbial metagenome when compared to wolves. Additional genes producing key enzymes in carbohydrate metabolic pathways were also more abundant in the dog microbial metagenome.[13]
A lack of the amino acid cysteine causes several diseases in many organisms, including dogs and wolves. Cysteine is found in many meat products, and carnivorous organisms usually receive enough cysteine through their diet. The dietary transition in dogs from raw meat to commercial dog food might cause a reduced intake of cysteine. The gut microbiome in dogs demonstrated an adaptation showing gut microbiota capable of synthesizing cysteine from cystathionine. Cystathionine can be derived from other amino acids found in foods. This demonstrates another pathway where bacteria can maintain cysteine levels in dogs to prevent deficiencies and illnesses.[14]
Conclusion
Dogs evolved from wolves through domestic interactions with humans. This most likely occurred between 11-35 kya in Central/Southeast Asia, the Middle East/Africa, and/or Europe. Subsequently, the change in diet caused a shift in the gut microbiome, allow dogs to more efficiently digest carbohydrates. There was also a shift in antibiotic resistance as an indirect byproduct of antibiotics used on farms and in the clinical setting.
References
- ↑ Yamashiro, K. (2023). Kuma (Photograph). Flickr.
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 2.16 2.17 Freedman AH, Wayne RK. Deciphering the origin of dogs: From fossils to genomes. Annual Review of Animal Biosciences. 2017 Feb 8;5(1):281-307
- ↑ Larson G, Karlsson EK, Perri A, Webster MT, Ho SY, Peters J, Stahl PW, Piper PJ, Lingaas F, Fredholm M, Comstock KE. Rethinking dog domestication by integrating genetics, archeology, and biogeography. Proceedings of the National Academy of Sciences. 2012 Jun 5;109(23):8878-83.
- ↑ Frantz LA, Mullin VE, Pionnier-Capitan M, Lebrasseur O, Ollivier M, Perri A, Linderholm A, Mattiangeli V, Teasdale MD, Dimopoulos EA, Tresset A. Genomic and archaeological evidence suggest a dual origin of domestic dogs. Science. 2016 Jun 3;352(6290):1228-31.
- ↑ 5.0 5.1 Vilà C, Savolainen P, Maldonado JE, Amorim IR, Rice JE, Honeycutt RL, Crandall KA, Lundeberg J, Wayne RK. Multiple and ancient origins of the domestic dog. Science. 1997 Jun 13;276(5319):1687-9.
- ↑ Savolainen P, Zhang YP, Luo J, Lundeberg J, Leitner T. Genetic evidence for an East Asian origin of domestic dogs. Science. 2002 Nov 22;298(5598):1610-3.
- ↑ 7.0 7.1 Pang JF, Kluetsch C, Zou XJ, Zhang AB, Luo LY, Angleby H, Ardalan A, Ekström C, Sköllermo A, Lundeberg J, Matsumura S. mtDNA data indicate a single origin for dogs south of Yangtze River, less than 16,300 years ago, from numerous wolves. Molecular biology and evolution. 2009 Dec 1;26(12):2849-64.
- ↑ Brown SK, Pedersen NC, Jafarishorijeh S, Bannasch DL, Ahrens KD, Wu JT, Okon M, Sacks BN. Phylogenetic distinctiveness of Middle Eastern and Southeast Asian village dog Y chromosomes illuminates dog origins. PLoS One. 2011 Dec 14;6(12):e28496.
- ↑ Boyko AR, Boyko RH, Boyko CM, Parker HG, Castelhano M, Corey L, Degenhardt JD, Auton A, Hedimbi M, Kityo R, Ostrander EA. Complex population structure in African village dogs and its implications for inferring dog domestication history. Proceedings of the National Academy of Sciences. 2009 Aug 18;106(33):13903-8.
- ↑ Vonholdt BM, Pollinger JP, Lohmueller KE, Han E, Parker HG, Quignon P, Degenhardt JD, Boyko AR, Earl DA, Auton A, Reynolds A. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature. 2010 Apr 8;464(7290):898-902.
- ↑ Thalmann O, Shapiro B, Cui P, Schuenemann VJ, Sawyer SK, Greenfield DL, Germonpré MB, Sablin MV, López-Giráldez F, Domingo-Roura X, Napierala H. Complete mitochondrial genomes of ancient canids suggest a European origin of domestic dogs. Science. 2013 Nov 15;342(6160):871-4.
- ↑ Skoglund P, Ersmark E, Palkopoulou E, Dalén L. Ancient wolf genome reveals an early divergence of domestic dog ancestors and admixture into high-latitude breeds. Current Biology. 2015 Jun 1;25(11):1515-9.
- ↑ 13.0 13.1 13.2 13.3 13.4 13.5 Liu Y, Liu B, Liu C, Hu Y, Liu C, Li X, Li X, Zhang X, Irwin DM, Wu Z, Chen Z. Differences in the gut microbiomes of dogs and wolves: roles of antibiotics and starch. BMC Veterinary Research. 2021 Dec;17:1-8.
- ↑ Lyu T, Liu G, Zhang H, Wang L, Zhou S, Dou H, Pang B, Sha W, Zhang H. Changes in feeding habits promoted the differentiation of the composition and function of gut microbiotas between domestic dogs (Canis lupus familiaris) and gray wolves (Canis lupus). Amb Express. 2018 Dec;8:1-2.
Edited by Roman Cicero, student of Joan Slonczewski for BIOL 116, 2024, Kenyon College.