HIV Envelope and Cell Fusion
Introduction
By Ian Perrone
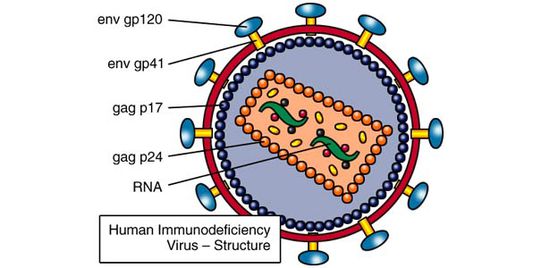
HIV, or human immunodeficiency virus, is an enveloped sense-RNA retrovirus that infects human immune cells in order to replicate (figure 1). Its classification as a retrovirus is due to its use of reverse transcriptase to copy its RNA genome into DNA. This DNA is imported into the host cell nucleus, where it is inserted into the host cell genome by virally encoded integrase. At this point, the virus lives as a prophage, sporadically producing infective virions through use of host transcriptase and ribosomes. Specifically, HIV is known as a lentivirus, a form of retrovirus with a long incubation time due to relatively slow replication. Despite this relatively slow replication speed, unless treated with HAART (highly active anti-retroviral therapy), the vast majority of HIV patients will eventually develop AIDS (acquired immunodeficiency syndrome). AIDS is characterized by extremely heightened susceptibility to opportunistic infection by rarely pathogenic bacteria, such as Pneumocystis carinii, and the development of uncommon types of cancer, such as Kaposi sarcoma [6]. Since the original spread of the virus from Africa to the USA in 1981, HIV and AIDS have caused the deaths of over 25 million people across the globe [6].
Like all enveloped viruses, HIV has envelope proteins that recognize particular protein structures on target cells. These envelope proteins are what allow viruses to bind to cells before fusing with the cell membrane entering the cytoplasm. These proteins also define the viral tropism, or range of cells that the virus can infect, making knowledge of the viral envelope proteins necessary for understanding the pathogenicity of the virus [2]. In addition, the key role played by the envelope proteins in infecting target cells means that a wide variety of treatments for viral diseases have been designed to block the action of these envelope proteins. The design of these binding/fusion blockers also requires a firm grasp of viral envelope proteins, especially in HIV, which has an especially high mutation rate due to its use of relatively inaccurate reverse transcriptase for replication. Below, we explore the structural characteristics of the main HIV envelope protein components, gp120 and gp41, as well as their respective targets. We also examine the influence these proteins have on viral evolution patterns, with a specific focus on the evolution of viral quasispecies within individual patients. Finally, we consider the potential for HIV treatments targeting these envelope proteins, as well as the difficulties associated with treatment of such a mutation-prone virus.
HIV Env Overview
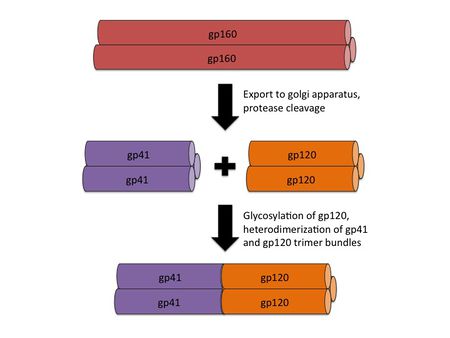
The envelope of the HIV virion consists of a glycoprotein complex, called Env, embedded in a host-sourced phospholipid membrane. Each virion includes approximately 15 Env glycoprotein complexes [6]. Env itself consists of trimers of noncovalently bound gp120 and gp41 subunits. During replication, the integrated prophage is transcribed, producing Env mRNA that is read by endoplasmic reticulum ribosomes to produce an 845-870 amino acid precursor polypeptide. This precursor is modified with the addition of asparagine linked, high mannose content sugar chains, yielding the intermediate glycoprotein gp160 [12]. The gp160 glycoprotein forms homotrimers before being exported to the golgi apparatus, where host proteases digest the glycoprotein complex, yielding gp120 and gp41 subunits which remain as homotrimers. The gp120 and gp41 trimer bundles are further modified through N-glycosylation. This glycosylation step alone contributes a great deal to the variability of the Env protein structure; gp120, for example, has around 24 potential N-glycosylation sites allowing for a wide variety of possible N-glycosylation combinations [7, 12].
After N-glycosylation, the gp120 trimer bundles form noncovalent bonds with gp41 trimer bundles, yielding gp41-gp120 bundle heterodimers (figure 2). Finally, the mature Env proteins are sent to the cell membrane and are embedded in virions budding from the cell. The high mutation rate of HIV means that mutations to these Env components are fairly common, resulting in a large number of progeny virions with Env glycoprotein complexes that fail to mature properly or simply fall out of the viral envelope [12]. Virions with these non-viable Env protein complexes are rendered incapable of infection. The sheer number of virions produced, however, guarantees that at least a small portion of the progeny virions will have viable Env protein complexes.
gp120 Structure and Role
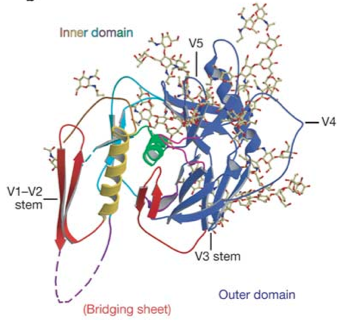
Env subunit gp120 serves as the target cell recognition and binding mechanism. Specifically, the outer domain of gp120 binds to the transmembrane protein CD4, which is found on the surface of CD4-expressing T helper cells, a key component of the human immune system. This interaction occurs within a largely hydrophobic binding pocket composed of β-ribbon components β20-β21, the CD4 binding loop, and the α-helices of the outer domain (see figure 3) [3, 12]. Binding to CD4 causes a series of conformational changes to occur in the inner domain of gp120 that bring together the β-sheets of the so-called "bridging sheet", which connects the outer and inner domains. This compacted bridging sheet serves as a binding site for the chemokine receptor CCR5, or less frequently, CXCR4 [3, 12]. Ultimately, these interactions serve to bring the virion closer to the target cell and cause the gp120 subunit to be shed from the viral envelope, exposing the gp41 subunit.
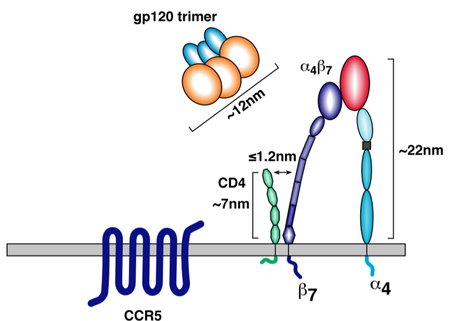
The gp120 subunit also features five variable loop regions known as V1, V2, V3, V4, and V5 [3, 9]. Some of these variable loops have been shown to directly interact with target cell membrane proteins important for productive infection. For example, the V1 and V2 loops are known to interact with the T-cell integrin α4β7 [5, 9]. This integrin is important for two reasons: first, it is found more frequently on mature T-cells that also express CD4 and CCR5, the HIV receptor proteins. In fact, α4β7 is usually found in the immediate vicinity of these two HIV receptor proteins (figure 4) [8, 9]. Thus, virions able to interact with α4β7 integrin have a better chance of binding to appropriate target cells. Secondly, α4β7 causes T-cell migration to the gut-associated lymphoid tissue (GALT), an important T-cell production center in the human gastrointestinal tract [5, 8]. The high density of CD4+ T-cells in this region allows for relatively easy intercellular transmission of the virus, making the GALT a hotbed of HIV activity. In cases of sexual transmission through the vaginal mucosa, α4β7 interaction allows HIV to hitch a ride to a region with much higher T-cell density, increasing the probability of productive infection [5, 8]. While interaction with α4β7 is not necessary for infection, it does improve the chances of establishing a productive infection [5, 8, 9]. Without V1 and V2, this important auxillary receptor interaction would be impossible.
The length of these variable loops fluctuates greatly due to the high mutation rate of HIV. In addition, the N-glycosylation state of the variable loops depends largely on mutations that add or remove potential N-glycosylation sites (PNGs) [9]. Together, the high variability in length and extent of glycosylation of the variable loops can have a profound impact on virion-target cell binding efficiency and specificity. For example, mutations to V3 can cause the virion to prefer the co-receptor CXCR4 to CCR5, and artificial deletion of V3 renders the virion incapable of binding to CCR5 at all [3]. In addition, mutations to V1 and V2 that remove PNGs have been shown to increase the efficiency of α4β7 interaction [9].
Interestingly, the high variability of the variable loops also serves as a mechanism for host immune system evasion. The V1 and V2 loops serve as frequent targets for autologous neutralizing antibodies produced in response to HIV infection [9]. The high mutation rate allows new generations of virus to escape these antibodies in a phenomenon called conformational masking. There are two mechanisms through which this masking is thought to occur: additional N-glycosylation of the V1 and V2 loops, and rearrangements of the existing V1 and V2 PNGs. These changes to the N-glycosylation pattern of V1 and V2 mask the viral Env protein, preventing recognition by neutralizing antibodies, which are produced targeting old V1/V2 sequences [9].
gp41 Structure and Role
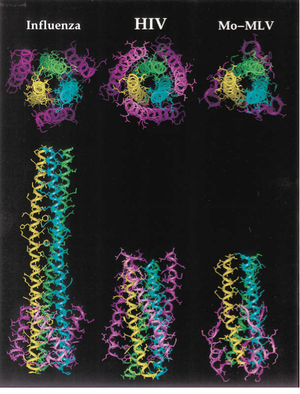
The gp41 subunit initiates fusion of the virion into the target cell after disassociation of gp120. The overall structure of gp41 includes of 3 domains: an ectodomain, a transmembrane domain, and a cytoplasmic domain [1, 13]. The gp41 trimer ectodomain consists of 3 parallel N-terminal α-helices in a coiled coil conformation, cradled by three antiparallel C-terminal α-helices connected by flexible loop linkers. The coiled coil ectodomain connects to the hydrophobic transmembrane domain, followed by the hydrophilic cytoplasmic domain. Interestingly, this structure is highly conserved amongst enveloped virus fusion proteins, bearing a striking resemblance to the influenza fusion protein HA2 in particular [1]. The diagram to the right compares structures of the trimer ectodomains from the enveloped viruses influenza, SIV (simian immunodeficiency virus, which is very closely related to HIV), and Mo-MLV (murine leukemia virus, a cancer-causing retrovirus) (Figure 5). Like influenza HA2, the coiled coil structure of gp41 is thought to act like a spring-loaded harpoon, projecting the so-called "fusion peptide" through the target cell membrane [1, 13]. The virion then proceeds to insert itself into the cell using gp41 as a tether. In the prefusogenic structure of gp41, this fusion peptide is obstructed by gp120. After gp120 binds to the HIV coreceptors and dissociates, however, the fusion peptide is exposed and conformational changes occur in the gp41 trimer that launch it into the target cell [1]. Besides serving as the cell fusion initiator, gp41 also mediates the processing of the gp160 intermediate, as well as the gp120/gp41 complex. Insertion and deletion mutations to the inner coiled coil motifs have been shown to prevent proper gp160 cleavage and gp120/gp41 complex formation [1, 13].
Env and Viral Evolution
As discussed above, the high mutation rate of HIV means that replication produces a large number of nonviable viral particles. The central role in infection played by the Env protein subunits means that mutations to the Env gene in the HIV genome frequently lead to crippled virions. At the same time, this high mutation rate is also responsible for conformational masking resulting from mutations in the V1 and V2 loops in the gp120 subunit. Crucially, the exact PNG additions or rearrangements to the V1 and V2 loops do not matter as long as they result in a structure that is sufficiently different from the structures of previous viral generations, which have been recognized and targeted by the host immune system [9]. The additional fact that these structural mutations to the gp120 subunit can have important effects on the coreceptor binding affinities of the virion means that these mutations produce a wide variety of viable virions with different degrees of vulnerability to the immune system and target cell binding preferences. Thus, instead of the virus existing as a single entity in a patient, replicating HIV is more like a large collection of related quasispecies, each with specific strengths and weaknesses. The existence of these viral quasispecies allows rapid evolution of the virus to changing conditions within the host, including the specificity of the latest batch of neutralizing antibodies [9].
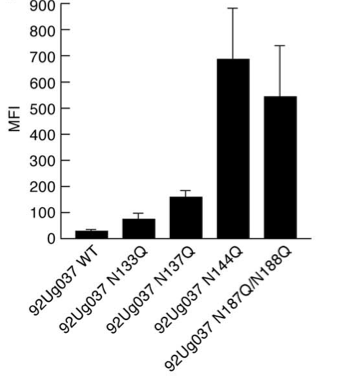
Frequently, these various selective pressures can cause changes to the structure of the Env protein subunits that seem to reduce the viability of the virion in other ways. Intriguingly, recent research shows that low V1 and V2 loop N-glycosylation increases the affinity of gp120 for α4β7 integrin, the gut-homing membrane protein introduced above [9]. Figure 5 shows data from a α4β7 binding assay in which PNGs in the V1 and V2 loops were removed by replacement of key asparagine residues with glutamines. This experiment shows that the removal of PNGs causes increased α4β7 binding affinity, with some PNG deletions contributing more than others. As we have just seen, however, increased N-glycosylation of the V1 and V2 loops is often associated with increased ability to evade the host immune system. Thus, this decreased vulnerability to the host immune system frequently reduces the efficiency of binding to α4β7 integrin, apparently resulting in reduced viability. Recent research suggests that while α4β7 binding affinity is important for establishment of productive infection, it is less important for virions produced in a host several months after the initial infection, as they are already replicating in tissue with high target cell density [9].
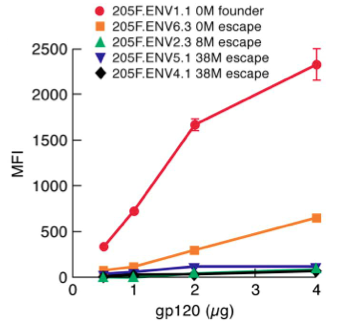
In addition, these late-stage virions are under the additional selective pressure of evading the host immune system. Together, the relative unimportance of α4β7 binding affinity and importance of evading neutralization by host antibodies for the viability of late-stage virions causes a trend of greater N-glycosylation of gp120 variable loops in clinical isolates from long-infected patients [9]. Thus, structural changes in Env are subject to selective pressures based on the stage of the infection, with low N-glycosylation, high α4β7 affinity quasispecies of HIV predominating early in the infection and relatively high N-glycosylation, immune evasion-capable quasispecies predominating late in the infection. Figure 6 to the right shows an experiment comparing the α4β7 binding affinity of early and late HIV clinical isolates from one female patient [9]. The isolates show α4β7 binding affinity (measured in terms of mean fluorescence intensity, MFI) that decreases as the time since the initial infection increases, an example of the increasing importance of immune system evasion as the infection progresses.
Due to the relatively high barriers to sexual transmission (receptive penile-vaginal unprotected sex involving an HIV+ male results in transmission of HIV less than 0.2% of the time, for example), the evolutionary pattern of HIV quasispecies resembles a shotgun blast: productive infection is commonly established by one or two quasispecies which probably have high α4β7 binding affinity, at which point a large number of quasispecies capable of host immune system evasion are able to differentiate from the founder quasispecies [5, 9]. Despite its relative vulnerability to neutralization by host antibodies, the initial high α4β7 affinity quasispecies are likely maintained in the viral population as prophages in infected T-cells, supplying good founder quasispecies for further infections. Alternatively, quasispecies with high N-glycosylation states that do not interfere with α4β7 interaction may develop, yielding high α4β7 affinity quasispecies that also feature good immune system evasion as was observed in one recent study [9]. This indicates that it is not simply N-glycosylation number that disrupts α4β7 interactions, but is the N-glycosylation of particular V1 or V2 loop sites that causes this decrease in binding affinity.
The pattern of HIV quasispecies evolution is complicated further by the characteristics of the cells predominantly infected by the virus. CD4+ T-cells come in a variety of subtypes, including central memory T-cells (TCM) and transitional memory T-cells (TTM). Recent research shows that patients treated with HAART soon after initial infection have a much higher ratio of TCM cells to TTM cells than patients without HAART treatment or patients put on HAART long after initial infection . In these early treatment patients, TCM cells make up the majority of the viral prophage reservoir, or the population of cells infected with latent HIV. While TCM cells can live for many years, partially explaining the difficulty of eradicating HIV even with HAART, they normally have low rates of homeostatic proliferation. Some TCM subtypes proliferate at a higher rate through antigen-mediated proliferation, however, leading to selection of the quasispecies of HIV that happen to have infected these high proliferation TCM cells [4]. In contrast, TTM cells proliferate more regularly through homeostatic proliferation, maintaining a viral reservoir of roughly consistent size and genetic diversity [4]. Thus, while the evolution of Env in HIV quasispecies is largely controlled by the stage of the infection, the point at which HAART treatment is initiated has important repercussions on the genetic diversity of the viral reservoir.
Env and HIV Treatment
Immunity-granting vaccines exist for a wide variety of pathogenic viruses. These viruses usually make use of native or modified virus envelope components, including fusion proteins. While a wide variety of HIV vaccines have been tested in clinical trials, only a few have been shown to offer any protection against infection, and this protection is partial at best. Unfortunately, the high variability of the Env components makes it extremely difficult to design a vaccine that covers all of the possible varieties of the Env subunits [6]. In other words, while a vaccine might protect against particular quasispecies of HIV, it fails to offer adequate protection against the many other quasispecies that can serve as founders of productive infection.
Despite this bad news, the crucial role of the Env subunits has lead to a great deal of interest in developing treatments for HIV that target some aspect of their function and limit or eliminate viral replication. While the various component drugs of HAART - such as reverse transcriptase, integrase, and viral protease inhibitors - usually do a good job of minimizing replication, the high mutation rate of HIV means that new treatments should be pursued to increase the diversity of therapy targets and so reduce the chances of drug resistance evolving [6]. Overall, there are two main approaches in the design of drugs targeting the function of the Env subunits: targeting the subunits themselves, and for gp120, targeting the HIV coreceptors CD4 and CCR5 or CXCR4 that initiate the binding and fusion process. While the first approach has yielded some potential fusion inhibitors designing compounds targeting the Env subunits themselves is difficult considering the high variability of Env subunit structure. The second approach has the advantage of targeting host cell membrane proteins with highly conserved structures. The initial discovery of the chemokine HIV coreceptors CCR5 and CXCR4 caused intense scrutiny of their natural ligands, such as MIP-1α/β for CCR5 and SDF-1α for CXCR4 with the aim of designing artificial ligands for these receptors that could block HIV binding or reduce coreceptor expresion [11]. The first concern in this design process was whether or not blocking these receptors caused serious problems in host immune system signaling. Individuals with double CCR5 knockout mutations exist in the human population without any known detrimental effects, granting these individuals relative immunity to R5-binding HIV and implying that CCR5 blockers are safe. Meanwhile, studies have shown that unblocked CXCR4 is important for normal development of fetal mouse models, making it difficult to design safe inhibitors for X4-binding HIV [11].
Initial studies developed modified versions of the natural CCR5 ligands that showed improved binding to the receptor in vitro. Unfortunately, these modified ligands were not stable enough to meaningfully block HIV infection in mouse models [11]. Subsequent studies have identified small-molecule inhibitors of HIV-coreceptor binding which have structures based on the natural ligands of CCR5 but are able to bind with higher affinity [11]. The first of these small-molecule inhibitors, TAK-779, was able to block HIV-coreceptor binding at nanomolar concentrations. Despite this, TAK-779 was not considered a viable therapeutic drug because it could not be taken orally and caused irritation when delivered by injection. Recently developed TAK-779 derivatives, such as TAK-220 and TAK-652, show promise as potential R5-binding HIV inhibitors, and can be taken orally without significant irritation [11].
Citations
1. Chan, D. C., Fass, D., Berger, J. M., & Kim, P. S. (1997). Core structure of gp41 from the HIV envelope glycoprotein. Cell, 89(2), 263-273. http://www.sciencedirect.com/science/article/pii/S0092867400802056
2. Chan, D. C., & Kim, P. S. (1998). HIV entry and its inhibition. Cell, 93(5), 681-684. http://cmgm3.stanford.edu/biochem/biochem230/papers2005/week3/Kim_Cell_1998Review.pdf
3. Chen, B., Vogan, E. M., Gong, H., Skehel, J. J., Wiley, D. C., & Harrison, S. C. (2005). Structure of an unliganded simian immunodeficiency virus gp120 core. Nature, 433(7028), 834-841. http://www.nature.com/nature/journal/v433/n7028/abs/nature03327.html
4. Chomont, N., El-Far, M., Ancuta, P., Trautmann, L., Procopio, F. A., Yassine-Diab, B., Boucher, G., Boulassel, M-R., Ghattas, G., Brenchley, J.M., Shacker, T.W., Hill, B.J., Douek, D.C., Routy, J-P., Haddad, E.K., & Sékaly, R. P. (2009). HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nature medicine, 15(8), 893-900. http://www.nature.com/nm/journal/v15/n8/abs/nm.1972.html
5. Cicala, C., Arthos, J., & Fauci, A. S. (2011). HIV-1 envelope, integrins and co-receptor use in mucosal transmission of HIV. J Transl Med, 9(Suppl 1), S2. http://www.biomedcentral.com/content/pdf/1479-5876-9-S1-S2.pdf
6. Girard, M. P., Osmanov, S., Assossou, O. M., & Kieny, M. P. (2011). Human immunodeficiency virus (HIV) immunopathogenesis and vaccine development: a review. Vaccine, 29(37), 6191-6218. http://www.sciencedirect.com/science/article/pii/S0264410X11009595
7. Leonard, C. K., Spellman, M. W., Riddle, L., Harris, R. J., Thomas, J. N., & Gregory, T. J. (1990). Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. Journal of Biological Chemistry, 265(18), 10373-10382. http://www.jbc.org/content/265/18/10373.short
8. McKinnon, L. R., Nyanga, B., Chege, D., Izulla, P., Kimani, M., Huibner, S., Gelmon, L., Block, K.E., Cicala, C., Anzala, A. O., Arthos, J., Kimani, J., & Kaul, R. (2011). Characterization of a human cervical CD4+ T cell subset coexpressing multiple markers of HIV susceptibility. The Journal of Immunology, 187(11), 6032-6042. http://www.jimmunol.org/content/187/11/6032.short
9. Nawaz, F., Cicala, C., Van Ryk, D., Block, K. E., Jelicic, K., McNally, J. P., Ogundare, O., Pascuccio, M., Patel, N., Wei, D., Fauci, A.S. & Arthos, J. (2011). The Genotype of Early-Transmitting HIV gp120s Promotes α4β7–Reactivity, Revealing α4β7+/CD4+ T cells As Key Targets in Mucosal Transmission. PLoS pathogens, 7(2), e1001301. http://www.plospathogens.org/article/info%3Adoi%2F10.1371%2Fjournal.ppat.1001301#ppat-1001301-g007
10. Pancera, M., Majeed, S., Ban, Y. E. A., Chen, L., Huang, C. C., Kong, L., Kwon, Y.D., Stuckey, J., Zhou, T., Robinson, J.E. Schief, W.R., Sodroski, J., Wyatt, R., & Kwong, P. D. (2010). Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proceedings of the National Academy of Sciences, 107(3), 1166-1171. http://www.pnas.org/content/107/3/1166.short
11. Ray, N., & Doms, R. W. (2006). HIV-1 coreceptors and their inhibitors. Chemokines and Viral Infection (pp. 97-120). Springer Berlin Heidelberg. http://link.springer.com/chapter/10.1007/978-3-540-33397-5_5#page-1
12. Wyatt, R., & Sodroski, J. (1998). The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science, 280(5371), 1884-1888. http://www.sciencemag.org/content/280/5371/1884.short
13. Yang, Z. N., Mueser, T. C., Kaufman, J., Stahl, S. J., Wingfield, P. T., & Hyde, C. C. (1999). The crystal structure of the SIV gp41 ectodomain at 1.47 Å resolution. Journal of structural biology, 126(2), 131-144. http://www.sciencedirect.com/science/article/pii/S1047847799941163