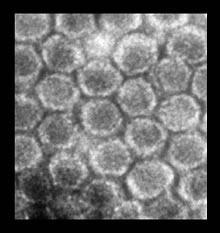
Helminthosporium victoriae Virus 190S
A Microbial Biorealm page on the genus Helminthosporium victoriae Virus 190S
Classification
Virus; dsRNA virus; Totiviridae; Victorivirus; Helminthosporium victoria Virus 190S
Description and Significance
Helminthosporium victoria virus 190S (HvV190S) is a virus of the victorivirus family that infects plant pathogenic fungi or protozoa [4]. It has been found to be the cause of Victoria blight in oats and is related to a lytic disease in the fungi it inhabits. HvV190S causes growth reduction, an increase in sectoring, and mycelial collapse [2]. HvV190S also triggers lysis, the degradation of a cell due to the rupture of the cellular membrane. Although this virus tends to be harmless to its host, HvV190S has been known to be highly debilitating depending on its particular fungal host and how it was raised [4]. The virus can have a hypovirulence or hypervirulence.
Viruses of phytopathogenic fungi have become increasing popular in experimental studies to determine and develop control measures that allow the host to withstand the virus rather than it having a fatal effect [4]. In addition, fungi has developed methods allowing adaptation to the virus and the ability to tolerate the virus’s replication in small doses by utilizing genes known as “super killers” that regulate the cope number of HvV190S to limit its growth [3].
Genome Structure
HvV190S has a linear, nonsegmented double stranded RNA (dsRNA) genome that’s between 4.6 and 6.3 kilo-base pairs in size [2]. The entire nucleotide sequence, 5178 base pairs, of HvV190S has been determined and is available online [4]. In addition to the dsRNA found in all HvV190S, some forms of the virus also contain satellite dsRNAs (encoding for “killer” proteins) that are separately surrounded by a protein shell [5].
The dsRNA consists of two long, open reading frames (ORFs) [6]. The upstream ORF, GAG, encodes for the capsid protein, and the downstream ORF, POL, encodes for the important RNA-dependent RNA polymerase (RdRp) [5]. The two ORFs overlap at the tetranucleotide AUGA region; therefore, the cell must have the ability to utilize both strands at the overlapping point [6]. Translation of the RdRp of the downstream ORF depends on the ability of the ribosome to reactivate and reinitiate after already completing termination from translation of the upstream ORF. In order for this to occur, studies have shown that a start-stop coupled termination-reinitiation strategy is practiced. A secondary structure called a pseudoknot (aka a slippery site) located at the overlap region within 32 nucleotides upstream of the AUGA region helps to slow down the movement of the ribosome unit to allow for the ribosome to undergo a frame shift and begin reinitiation. A frameshift is when the ribosome jumps from one reading frame to the next on an mRNA strand to allow for protein synthesis from the area of overlapping frames. According to studies, the close proximity of the pseudoknot and the stop codon of the capsid protein ORF plays an important role in the promotion of the downstream ORF’s translation process.
Cell Structure, Metabolism & Life Cycle
HvV190S comes in the form of virions. A virion is a non-enveloped particle 40 nm in diameter [5]. It has a relatively smooth outer coat called a capsid that is single-shelled and surrounds the single major polypeptide of genetic information [3]. The capsid has icosahedral symmetry and is composed of 120 capsid protein (CP) subunits having molecular masses between 76 and 98 kDa. It also consists of 60 asymmetric CP dimers. Two CP subunits exist; the A-subunits, which tend to cluster at the fivefold axis of the virion particle, and the B-subunits, which cluster near the threefold axis. The capsid also contains polypeptides p78, p83, and p88 [4]. P83 and p88 are both phosphoproteins, while p79 is nonphosphorylated. Virions of any particular virus can be identified due two differences in sedimentation rates as well as the varying compositions of these capsid polypeptides. HvV190S in particular is a mixture of two different capsid types: 190S-1 and 190S-2. 190S-1 is composed of equimolar amounts of p88 and p83, and 190S-2 is composed of p88 and p78 is similar quantities.
The victorivirus family is a newly identified family of the totivirus genus. Not much is known yet about the metabolic process and constituents of family victorivirus besides that they utilize the nutrients of their host for survival and growth [4].
Replication of HvV190S is intracellular [4]. The virus undergoes a conservative process where the RNA polymerase performs transcription from end-to-end of the dsRNA strand in a catalytic manner [3]. This newly synthesized strand becomes the template strand for translation [5]. The (+) RNAs are encapsidated in a newly synthesized virion particle where it is transcribed to form (–) RNA molecules that are then base paired and soon form the new dsRNA genome. Once the new genome has been formed, the mature virion particle is distributed to new cells through cell division, spore formation, or even cell fusion.
Ecology (including pathogenesis)
HvV190S inhabits plant pathogenic fungi and protozoa [4]. Pathogenic means that these fungi are able to cause disease. The virus is typically not harmful to its host, but on occasion can sometimes be damaging. HvV190S causes disease by contaminating its host, which in turn affects the plants on which is grows. HvV190S was first discovered to be the reason of Victoria blight disease in oats. It causes root rotting, a stunt in seed formation, and color streaking on leaves, often ultimately leading to plant death [1]. Other virulence factors of HvV190S that may actually affect the host fungi are decrease in growth, an increase in sectoring, and mycelial collapse, a breakdown of the vegetative region of the fungi or plant [2].
Interesting Feature
A particularly interesting effect HvV190S has on plants is that it causes them to produce antifungal proteins as an attack mechanism against the HvV190S-infected fungal hosts [2]. Some common antifungal proteins include killer proteins, defensins, and PR-1 proteins, along with many others. However, a newly discovered antifungal protein, victoriocin, found in plants infected by HvV190S, acts similarly to killer proteins. Victoriocin is encoded by the vin gene, located in the host. A study was done showing that virus-free host strains had almost no antifungal protein activity, suggesting that increased expression of the vin gene, and therefore increased production of victoriocin, would only be present in HvV190S-infected hosts. This was proven when overexpression of the vin gene was induced in a virus-free strain and the host showed increased production of victoriocin to order to fight the virus that wasn’t really there. The complexity of internal plant mechanisms is outstanding. Plants have the ability to fight against viruses by producing antifungal proteins that work to kill of the HvV190S-infected fungi.
References
1. Christensen, Jane. "Victoria Blight of Oats." University of Nebraska-Lincoln Department of Plant Pathology. J.E. Partridge, 2008. Web. 30 Oct. 2011. <http://nu-distance.unl.edu/homer/Disease/agron/cereals/OtVicBlt.html>.
2. De Sá, Patricia B., Wendy M. Havens, and Said A. Ghabrial. "Characterization of a Novel Broad-Spectrum Antifungal Protein from Virus-Infected Helminthosporium (Cochliobolus) Victoriae." Phytopathology 100.9 (2010): 880-89. APS Journal. The American Phytopathological Society. Web. 30 Oct. 2011. <http://apsjournals.apsnet.org/doi/pdf/10.1094/PHYTO-100-9-0880>.
3. Ghabrial, Said A. "Totiviruses." Desk Encyclopedia of Plant and Fungal Virology. By Brian W.J. Mahy and Marc H.V. Van Regenmortel. Oxford: Academic, 2010. 565-76. Print.
4. Huang, Shaohua, and Said A. Ghabrial. "Organization and Expression of the Double-stranded RNA Genome of Helminthosporium Victoriae 190S Virus, a Totivirus Infecting a Plant Pathogenic Filamentous Fungus." Proceedings of the National Academy of Sciences 93.22 (Oct. 1996): 12541-46. National Center for Biotechnology Information. Stanford University's Highwire Press. Web. 30 Oct. 2011. <http://www.ncbi.nlm.nih.gov/pmc/articles/PMC38028/pdf/pnas01526-0499.pdf>.
5. Hulo, C., E. De Castro, and P. Masson Et Al. "Victorivirus." 39 (2011): D576-82. ViralZone. Swiss Institute of Bioinformatics, Jan. 2011. Web. 30 Oct. 2011. <http://viralzone.expasy.org/all_by_species/745.html>.
6. Li, Hua, Wendy M. Havens, Max L. Nibert, and Said A. Ghabrial. "RNA Sequence Determinants of a Coupled Termination-Reinitiation Strategy for Downstream Open Reading Frame Translation in Helminthosporium Victoriae Virus 190S and Other Victoriviruses (Family Totiviridae)." Journal of Virology 85.14 (July 2011): 7343-37352. American Society for Microbiology. Web. 30 Oct. 2011. <http://jvi.asm.org/cgi/content/abstract/85/14/7343>.