Interactions between commensal bacteria and plant immune systems
Introduction
Commensal bacteria are a prominent and diverse community of bacteria that act on their host’s immune system in order to induce responses that prevent invasion and growth of pathogens. In humans, commensal bacteria often inhabit mucous membranes and epidermal surfaces. They also play an integral role in helping to protect their host from respiratory pathogens[1] . In addition to preventing pathogens from colonizing their host, commensal bacteria are also able to help regulate the immune system, and educate it in distinguishing pathogenic bacteria[2] . There has been a lot of research done on the diverse yet crucial roles that the trillions of commensal bacteria in humans play. There exist diverse and large communities of these bacteria in plants as well. However, their role, transmission, and interactions with the plants, in particular their immune system, is still being studied. In plants, much research is being done in how these bacteria interact with the root immune system, as they seem to play a large role in protecting the plant from pathogens, despite many containing the microbe associated molecular patterns(MAMPs) that should elicit an immune response from the host. Further research is also being performed on topics such as host preference, bacterial response to environmental stressors, and what genetic components allow the commensal bacteria to protect the host plant.
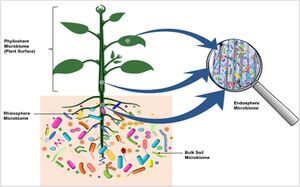
Microbiome
The plant microbiome is a community of diverse microorganisms that occupy the endosphere and episphere of plant tissue[3] . These communities play integral roles in the health, ecology, and productivity of the host plant. The endosphere refers to the area inside of plant tissue[4] . However, there are also populations of bacteria that are found in the rhizosphere and phyllosphere, which refer to the roots and soil, and stems and leaves respectively, as shown in Figure 1. All three of these environments contain large and markedly different microbiota, including the phyllosphere which is notoriously hostile for microorganisms. In the phyllosphere, there is little in the way of nutrients and water availability is inconsistent, yet nonetheless there is a complex population of microbes residing there[5] . Typically, the microorganisms in the plant microbiome are acquired from the environment. Microbes in the soil are attracted and enriched by chemotaxis and nutrients from the roots, and go on to colonize the plants as a result[6] . The plant microbiome plays an important role in keeping the plant healthy, as not only do the commensal bacteria compete with pathogens for things like nutrients and space, but they also directly produce microbicidal agents that aid the plant immune system in defending the plant from invaders[7].
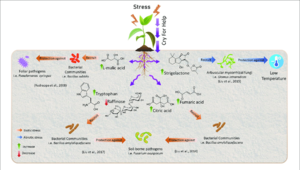
Response to Stress
One important distinction to make in regards to the plant microbiome, is that it is not a static entity. As the environment and stressors around the plant host change, so too will the composition and function of the microbiome[8] . While it was once thought that the changes in the microbiome were simply passive responses of plants, recent studies are suggesting that this may not be the case. Evidence indicates that as a result of many years of coevolution, plants actively seek the help of microbes in order to combat stress[9]. Some studies demonstrated that plants employ a strategy termed “cry for help”, where after experiencing stress from things like pathogens, plants can use chemical stimuli to recruit additional commensal bacteria in order to combat the stresses[10],as shown in Figure 2 .One recent experiment showed that when exposed to a fungal pathogen, sugar beet roots would recruit commensal bacteria such as Chitinophaga and Flavobacterium in order to help combat and suppress the pathogen[11]. Other studies have demonstrated that exposure of plants to stress can result in plants picking up beneficial microbes that go on to be inherited in future generations[12]. Many recent studies have investigated the ability of microbes to rescue plants from stresses, and a number of different strains of bacteria, such as Chitinophaga spp.,Chryseobacterium spp., Flavobacterium spp., Microbacterium spp.,Pseudomonas spp.,Sphingomonas spp.,Stenotrophomonas spp., and Xanthomonas spp have been observed enriching plants as a result of attacks from pathogens[13].
Genetics
Suppressing plant immune system
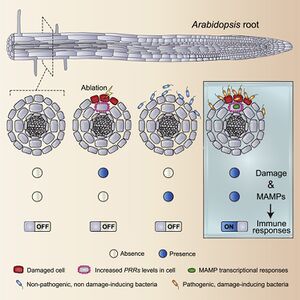
In a given plant, there is often a multitude of commensal bacterias that have beneficial effects on the host’s health[14]. In order to combat invasive pathogens, the plant immune system implements biochemical responses after recognizing a microbial molecule based on its microbe-associated molecular pattern, or MAMP, in a process called MAMP triggered immunity. The commensal bacteria that reside in and aid the host plants, often have the same MAMPs that would trigger an immune response against a harmful pathogen[15].Many studies have been performed in an attempt to understand why, despite having these patterns, commensal microbes don’t elicit the same immune response as other pathogens. Recent studies have suggested that commensal bacteria have evolved the ability to suppress MAMP triggered immunity, also known as MTI. One experiment found that commensal strains such as P. simiae WCS417 and Bacillus subtilis FB17 could suppress MAMP response genes in arabidopsis roots[16]. This supports the findings of other studies, which have suggested that commensal microbes can interfere with specific parts of the plant immune response.[17]Furthermore, studies indicate that the ability to suppress the host plant’s immune system has actually evolved multiple times in commensal bacteria, suggesting that there are numerous mechanisms for suppressing immune response in the bacteria[17].Other studies have suggested that it is because commensal bacteria avoid damage to the cell, a signal that enhances immune response in root tissues as shown in Figure 3, that they are able to colonize the plant[18].There is still much that is being studied in how commensals are able to colonize plants, however, it does appear that there is evidence to suggest that their ability to interfere with MTI stems primarily from nonspecific strategies, as commensals seem to express a much lower degree of host specification[17].
Host Preference
Even among plant species that live in identical environments, the roots are oftentimes colonized by entirely distinct communities of bacteria[19]. One study compared communities across very different land plants, and found that there was a separation by host plant species despite there being conversion among higher ranks of taxonomy[20].Despite extensive research, it is still not entirely known whether certain commensal are simply adapted to a specific plant species, or if plants are able to actively select for a certain type of commensal[19].However, there is still much research being performed in this area, such as one study that looked to investigate host preference of commensal communities. The results of this study suggested that commensals adapt to host features based on their respective genera.[19]However, other studies have suggested that instead, host preference patterns were based on ecological fitting, and the ability of a bacteria to colonize and survive with their already possessed traits[21].
Pseudomonas
One of the most commonly studied plant commensal bacteria is Pseudomonas. These are bacteria that typically inhabit the rhizosphere, and aid the host plant immune system by producing metabolites, such as antibiotics that are toxic to common plant pathogens[22]. One experiment sequenced the genome of a Pseudomonas strain called Pseudomonas Fluorescens 5, or PF5, in order to identify features that contributed to its commensal nature[23].This strain in particular inhabits the rhizosphere of a multitude of plants, and aids in suppressing diseases caused by soilborne pathogens[24]. Recent studies have found that PF5 is capable of producing a wide variety of antibiotics such as pyrrolnitrin, pyoluteorin and 2,4-diacetylphloroglucinol[25].The diversity and capability of pseudomonas, has led to them being one of the most oft studied bacterial strains, and they often serve as a sort of control organism for rhizosphere competence[23]. Another study looked at a different strain of pseudomonas, and found that in each lineage different bacterial genes are linked to protecting the plant[26].The same study also found that among different lineages of pseudomonas, the function of plant protection remained constant, but that its form differed starkly even among closely related lineages[26].Results similar to these were found in a separate experiment, which focused on commensal bacteria that combats Ralstonia solanacearum, a pathogen in tomatoes[27].Recent evidence suggests that one of the most important ways that these bacteria help eliminate pathogens, beyond simply producing antibiotics, is by outcompeting pathogenic pseudomonas in iron uptake, a micronutrient which is typically quite limited in plants, and thus outcompeting the pathogens allows the commensal pseudomonas to help protect the host[28].
Conclusion
The health of a wide multitude of plants is highly dependent on the commensal bacteria community that the host supports. Beyond outcompeting pathogens and producing antibiotics, many studies have shown that changes in the root microbiome often have an impact on the stress tolerance of plants. Thus, regulating interactions between the microbiome and plant immune systems, are crucial for maintaining a strong stress tolerance. There is still much research that is being done into these bacterial communities, as there aren’t clear answers in regards to their ability to avoid triggering MTI and the determination of host preference, however the last decade alone has seen a noticeable uptick in the amount of research being conducted in this topic, as it is suggested that understanding these bacteria may have more far reaching implications for agricultural production optimization.
References
- ↑ Dastogeer, Khondoker MG, Farzana Haque Tumpa, Afruja Sultana, Mst Arjina Akter, and Anindita Chakraborty. "Plant microbiome–an account of the factors that shape community composition and diversity." Current Plant Biology 23 (2020): 100161.
- ↑ Hanning, Irene, and Sandra Diaz-Sanchez. "The functionality of the gastrointestinal microbiome in non-human animals." Microbiome 3, no. 1 (2015): 1-11.
- ↑ Dastogeer, Khondoker MG, Farzana Haque Tumpa, Afruja Sultana, Mst Arjina Akter, and Anindita Chakraborty. "Plant microbiome–an account of the factors that shape community composition and diversity." Current Plant Biology 23 (2020): 100161.
- ↑ Hardoim, Pablo R., Leo S. van Overbeek, and Jan Dirk van Elsas. "Properties of bacterial endophytes and their proposed role in plant growth." Trends in microbiology 16, no. 10 (2008): 463-471.
- ↑ Lindow, Steven E., and Johan HJ Leveau. "Phyllosphere microbiology." Current Opinion in Biotechnology 13, no. 3 (2002): 238-243.
- ↑ Ramírez-Puebla, Shamayim T., Luis E. Servín-Garcidueñas, Berenice Jiménez-Marín, Luis M. Bolaños, Mónica Rosenblueth, Julio Martínez, Marco Antonio Rogel, Ernesto Ormeño-Orrillo, and Esperanza Martínez-Romero. "Gut and root microbiota commonalities." Applied and environmental microbiology 79, no. 1 (2013): 2-9.
- ↑ Berlec, Aleš. "Novel techniques and findings in the study of plant microbiota: search for plant probiotics." Plant Science 193 (2012): 96-102.
- ↑ Liu, Hongwei, Laura E. Brettell, Zhiguang Qiu, and Brajesh K. Singh. "Microbiome-mediated stress resistance in plants." Trends in Plant Science 25, no. 8 (2020): 733-743.
- ↑ Liu, Hongwei, and Laura E. Brettell. "Plant defense by VOC-induced microbial priming." Trends in plant science 24, no. 3 (2019): 187-189.
- ↑ Bakker, Peter AHM, Corné MJ Pieterse, Ronnie de Jonge, and Roeland L. Berendsen. "The soil-borne legacy." Cell 172, no. 6 (2018): 1178-1180.
- ↑ Carrión, Víctor J., Juan Perez-Jaramillo, Viviane Cordovez, Vittorio Tracanna, Mattias De Hollander, Daniel Ruiz-Buck, Lucas W. Mendes et al. "Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome." Science 366, no. 6465 (2019): 606-612.
- ↑ Yuan, Jun, Jun Zhao, Tao Wen, Mengli Zhao, Rong Li, Pim Goossens, Qiwei Huang et al. "Root exudates drive the soil-borne legacy of aboveground pathogen infection." Microbiome 6, no. 1 (2018): 1-12.
- ↑ Lau, Jennifer A., and Jay T. Lennon. "Rapid responses of soil microorganisms improve plant fitness in novel environments." Proceedings of the National Academy of Sciences 109, no. 35 (2012): 14058-14062.
- ↑ Fitzpatrick, Connor R., Isai Salas-González, Jonathan M. Conway, Omri M. Finkel, Sarah Gilbert, Dor Russ, Paulo José Pereira Lima Teixeira, and Jeffery L. Dangl. "The plant microbiome: from ecology to reductionism and beyond." Annual review of microbiology 74 (2020): 81-100.
- ↑ Teixeira, Paulo José PL, Nicholas R. Colaianni, Connor R. Fitzpatrick, and Jeffery L. Dangl. "Beyond pathogens: microbiota interactions with the plant immune system." Current opinion in microbiology 49 (2019): 7-17.
- ↑ Lakshmanan, Venkatachalam, Sherry L. Kitto, Jeffrey L. Caplan, Yi-Huang Hsueh, Daniel B. Kearns, Yu-Sung Wu, and Harsh P. Bais. "Microbe-associated molecular patterns-triggered root responses mediate beneficial rhizobacterial recruitment in Arabidopsis." Plant Physiology 160, no. 3 (2012): 1642-1661.
- ↑ 17.0 17.1 17.2 Teixeira, Paulo JPL, Nicholas R. Colaianni, Theresa F. Law, Jonathan M. Conway, Sarah Gilbert, Haofan Li, Isai Salas-González et al. "Specific modulation of the root immune system by a community of commensal bacteria." Proceedings of the National Academy of Sciences 118, no. 16 (2021): e2100678118.
- ↑ Zhou, Feng, Aurélia Emonet, Valérie Dénervaud Tendon, Peter Marhavy, Dousheng Wu, Thomas Lahaye, and Niko Geldner. "Co-incidence of damage and microbial patterns controls localized immune responses in roots." Cell 180, no. 3 (2020): 440-453.
- ↑ 19.0 19.1 19.2 Wippel, Kathrin, Ke Tao, Yulong Niu, Rafal Zgadzaj, Niklas Kiel, Rui Guan, Eik Dahms et al. "Host preference and invasiveness of commensal bacteria in the Lotus and Arabidopsis root microbiota." Nature microbiology 6, no. 9 (2021): 1150-1162.
- ↑ Fitzpatrick, Connor R., Julia Copeland, Pauline W. Wang, David S. Guttman, Peter M. Kotanen, and Marc TJ Johnson. "Assembly and ecological function of the root microbiome across angiosperm plant species." Proceedings of the National Academy of Sciences 115, no. 6 (2018): E1157-E1165.
- ↑ Agosta, Salvatore J., and Jeffrey A. Klemens. "Ecological fitting by phenotypically flexible genotypes: implications for species associations, community assembly and evolution." Ecology Letters 11, no. 11 (2008): 1123-1134.
- ↑ Haas, Dieter, and Christoph Keel. "Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease." Annual review of phytopathology 41 (2003): 117.
- ↑ 23.0 23.1 Paulsen, Ian T., Caroline M. Press, Jacques Ravel, Donald Y. Kobayashi, Garry SA Myers, Dmitri V. Mavrodi, Robert T. DeBoy et al. "Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5." Nature biotechnology 23, no. 7 (2005): 873-878.
- ↑ Howell, C. R., and R. D. Stipanovic. "Control of Rhizoctonia solani on cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium." Phytopathology 69, no. 5 (1979): 480-482.
- ↑ Nowak-Thompson, Brian, Steven J. Gould, Jennifer Kraus, and Joyce E. Loper. "Production of 2, 4-diacetylphloroglucinol by the biocontrol agent Pseudomonas fluorescens Pf-5." Canadian Journal of Microbiology 40, no. 12 (1994): 1064-1066.
- ↑ 26.0 26.1 Shalev, Or, Haim Ashkenazy, Manuela Neumann, and Detlef Weigel. "Commensal Pseudomonas protect Arabidopsis thaliana from a coexisting pathogen via multiple lineage-dependent mechanisms." The ISME journal 16, no. 5 (2022): 1235-1244.
- ↑ Gu, Shaohua, Zhong Wei, Zhengying Shao, Ville-Petri Friman, Kehao Cao, Tianjie Yang, Jos Kramer et al. "Competition for iron drives phytopathogen control by natural rhizosphere microbiomes." Nature Microbiology 5, no. 8 (2020): 1002-1010.
- ↑ Butaitė, Elena, Michael Baumgartner, Stefan Wyder, and Rolf Kümmerli. "Siderophore cheating and cheating resistance shape competition for iron in soil and freshwater Pseudomonas communities." Nature communications 8, no. 1 (2017): 1-12.
Edited by Ethan Liu, student of Joan Slonczewski for BIOL 116 Information in Living Systems, 2022, Kenyon College.
