Megavirales Genetics from Mimivirus to Chilensis
By Adama Berndt
Introduction
The evolutionary history of viruses is an unclear and frequent topic of speculation. Megaviruses are especially unique, as their size and structure are reminiscent of bacterial cells, but their replicative functions are similar to those of traditionally smaller viruses[1]. Although knowledge of large viruses dates back to the 1970’s, interest in megaviruses expanded during 2003 with the sequencing of Mimivirus; now one of the most well characterized Nucleocytoplasmic Large DNA Viruses(NCLDVs).[2] There are 6 official recognized classes of NCLDVs: Poxviridae, Asfarviridae, Iridoviridae, Ascoviridae, Phycoviridae, Mimiviridae(Fig 2.).[3] In addition to these classes Megaviridae is proposed as a seventh class.[4] One of the most remarkable recurring traits of these microbes is their large genome size, which at approximately one million base pairs is bigger than some bacterial genomes. These base pairs code for over a thousand genes and this genetic material is surrounded by pseudo-icosahedral protein capsids, similar to a Herpes virus(Fig. 1.).[5] The taxonomy of viruses as DNA containing microbes has long been considered from many angles. Of special consideration is the status of NCLDVs as living or non-living organisms. From an evolutionary perspective, there are two major sides to this debate, the “virus-early” hypothesis and the “virus-late’ hypothesis. The virus-early argument consists of the idea that viruses originated before modern cells and the virus-late ideology is characterized by the idea that viruses are the result of a reductive evolution, due to increased benefit from reliance on host metabolism, which propelled the loss of primary cellular machinery. Anyone in the virus-late camp also might theorize a scenario where the escape of proteins and genetic information from cells coupled to form viruses.[6] If viruses were formed spontaneously from released cell genetic materials and proteins, we might expect to see some formation of new rudimentary viruses today. The genetic information obtained from Mimivirus has provided extra fuel for speculation in this debate, specifically that Mimivirus contains the first evidence of a virus containing genes for transcription and translation.[7] The full phylogenetic analysis conducted by Raoult and colleagues found several Eukaryotic protein homologues in Mimivirus, and support for Mimivirus as a sister group to Eukaryotes and therefore a new branch of life known as Megavirales that would stand separately from Eukarya, Prokarya and Archaea.
The Evolution of Nucleocytoplasmic Large DNA Viruses
One of the most fascinating aspects of NCLDVs, is how knowledge of their existence has changed the debate on viruses as life or not. NCLDVs share genes with all of the primary living domains; Archaea, Bacteria and Eukarya. It is especially integral that these genes are involved with the something as essential to life as DNA storage and processing.[8] It begs the question, how can these viruses contain so many genes required for life, and still not be considered life? In addition, it has recently been demonstrated that Mimivirus contains its own glycosylation that leads to a 4-amino-4 6-dideoxy-D-glucose, called viosamine. It is also possible that this pathway could have evolved and changed to become similar eukaryotic glycosylation pathways. The presence of this sugar is observed even without the presence of regular amoeba host, suggesting the potential for autonomous, or semi-autonomous growth of Mimivirus.[9] Environmentally the role of aquatic NCLDVs has also been imagined. With the suggestion being that Mimiviruses may have lysed prokaryotes or eukaryotes, causing the of release carbon containing compounds, or other nutrients. These nutrients then were responsible for feeding the presence of other microbes and animals. When it comes to considering virus-early or virus-late hypotheses, unfortunately there is not a clear cut answer in differing phylogenetic trees. Analysis of highly conserved genes suggest that both hypotheses are possible, eukaryotes may share an ancestor for all NCLDVs, until this ancestor eventually diverged from eukaryotic life. It is also possible that eukaryotes were already a distinguished group and played host to the ancestor of modern day NCLDVs, until their differentiation was mediated by horizontal virus transfer.[10]
Support For Megavirales as a Fourth Domain
The classification of NCLDVs as a defining phylogenetic group, or the establishment of Megavirales as a proposed new order is one that does not go unchallenged in the face of certain objections to phylogenies created from viral DNA. This is for two main reasons: Viruses evolve at an extremely rapid rate meaning that consistency will be difficult to maintain from one genetic analysis to the next, within a small set of constantly changing genes. The second reason is that viruses are liable to engage regularly in horizontal gene transfer with the host. Frequent gene transfer obscures clear relationships between species or patterns of descent. However, in the case of NCLDVs these objections are less applicable. These double-stranded DNA viruses do not evolve at a rate comparable to smaller viruses with homologs that are significantly conserved, therefore we might expect the significant difference between NCLDVs to be a result of divergence that has occurred over a significant amount of time, suggesting a common ancestor for NCLDVs. The idea that horizontal gene transfer occurs regularly is also challenged in Megaviruses, due to their ability to rely completely on the cellular processes of the host. Therefore, there is not much evolutionary incentive for them to retain host genomes in NCLDV populations, in fact the pressure would suggest more static gene populations, or further reductive evolution, barring some unforeseen environmental shift. There is also not genetic evidence for clear-cut examples of gene transfer that has occurred between Eukaryotic hosts and their associated NCLDVs. For example, the open reading frames for non-homologous genes between Megavirus and MImivirus are 80% different, much higher than would be expected in the case of horizontal gene transfer. However, certain sets of genes (described further in gene product section) are sufficiently conserved in order to facilitate and form an adequate phylogeny.[11] To build a phylogeny, the Legendre paper used two specific genes that had excellent conservation in NCLDV’s and that were necessary for DNA replication. The first being a ribonucleoside diphosphate reductase sub-unit and a DNA replication clamp loader called factor C. This analysis illustrates that these genes show clear distinction from Archaea and Eukarya, showing little, or no overlapping affinities between these orders. Said another way, Megavirales as a proposed order, appears just as separate from Eukarya as Archaea are. Also up for speculation is whether or not Megavirales are at the root of the Eukaryotic domain as in figure 3A, or are not evolutionary pre-cursors to eukaryotes as in figure 3B.
However, phylogeny based on these two gene regions as in figure 3. are not necessarily the only kind a researcher could choose to build. In fact other work using DNA ligase as the primary gene to build phylogeny, has not helped to demonstrate such a clear cut relationship.[12] When using DNA ligase Yutin and Koonin found that a presence-absence analysis of a particular kind of DNA ligase feautres clear progression and separation of an ancestral and current form. However, when supplementing the very same analysis with other DNA ligase genes, the likely ancestor became the more recent ligase and the other ligase was projected as the ancestor. This shows that although there is obvious support for a common ancestor for NCLDVs, not all phylogenies will have genes chronologically in the same order. Some phylogenies do not have statistical separation or grouping of the proposed NCLDVs with significance. It all depends on the genes you choose to analyze, and weighting certain genes as more or less important is extremely complicated and difficult.[13] Ultimately, some researchers reach the opposite conclusions contained in other papers. Specifically they hint at the possibility of independent acquisition of genes by NCLDVs from cellular hosts, as a way of explaining difficulties in consistently classifying these different large viruses. Support for Megavirales as a new life classification is also slightly complicated when considering that Poxviridae and Ascoviridae, are both NCLDVs that don’t feature icosahedral capsids, instead they have brick-shaped capsids. However, in Poxviridae organisms it is thought that this feature is a derived one, due to the fact that during morphogenesis the icosahedral form is expressed as an intermediate form, as opposed to the dominant form. However, the D13 protein is still transcribed and translated during this developmental stage.[14]
Sequencing Methods[15]

Roche-454 Pyrosequencing was one of the first methods to gain commercial use for rapid genetic sequencing and is in common use for obtaining the phylogeny analyses discussed earlier. Pyrosequencing is powered by the release of pyrophosphates at every nucleotide addition. This pyrophosphate release stimulates a cascade that results in the eventual light production by firefly enzyme luciferase. The measured light correlates to number of base pairs that have been constructed. (Mardis, 2008). First DNA is separated to form an ssDNA library by way of bonding to a multitude of agarose beads containing complementary oligonucleotide sequences. Then these beads are separated in mixtures of oil and water that isolate each bead in micelles with PCR reactants. This means that each bead ends up with millions of copies of a single strand of DNA. Each bead is then arranged and fixed in place by the wells of a picotiter plate. Agarose beads are then surrounded by enzyme-containing beads that are responsible for catalyzing associated reactions that facilitate nucleotide addition through the creation of downstream pathways. The isolated addition in all wells is read simultaneously by CCD cameras, as pure nucleotide solution is added. The emitted light allows the camera to tell which nucleotides are added and computer analysis may then align complementary strands to generate a whole sequence. This process affords 250 nucleotides per read, and the computer eliminates the presence of mixed beads with mixed initial ssDNA’s, or beads with DNA that is not affixed and is lacking the calibratory TCGA sequence. This system is not perfect however, due to the manner in which pyrosequencing is calibrated, reads of the same base over 6 times lead to a particular kind of error, although other errors are certainly possible, this is where other methods of sequencing can fill in the gaps.
The other method often used in conjunction with Roche 454 pyrosequencing is a combination of Illumina and Sanger techniques(Arslan, 2011). For Illumina sequencing DNA is transected randomly and bound to Illumina adapter sequences at each end of DNA. These adapter sequences tether to oligonucleotide flow cells and complementary adapter strands bind together to form bridges that facilitate repeated amplification of DNA at flow cell clusters. Eventually strands are replicated in clusters of optimally a million identical DNA strands similar to the Roche agarose clusters and then they are separated. Then to finally sequence the data the addition of differential ACGT fluorescent nucleotides is imaged after each unique fluorescent base binds one at a time. Then the data is analyzed for read quality, and any bad reads are discarded. Random sequences of 25-35 base pairs are assessed for similarity and overlapped according to repeat sequences. This level of automation constructed in sequencing genetic information is takes up large amounts of data storage and requires a relatively powerful computer for the sequencing software algorithms, but the time and money saved as a result as revolutionized the research that can be done in genetic analysis. It is now possible to gain detailed and potentially fruitful information about the proteins coded for by the isolates of novel microbe genomes, such as those of NCLDVs.
Mimivirus Replication
Acanthamoeba polyphaga Mimivirus is one of the most well studied of the Nucleocytoplasmic Large DNA Viruses (NCLDVs), and therefore serves as a good focusing organism when understanding NCLDV replicative cycles. To begin replication Mimivirus first must enter a host, which is usually an Amoeba, although some evidence suggests that Mimivirus has had interaction with coral species. [16] After entering the host, Mimivirus must undergo a 4 hour eclipse phase, this relatively inactive period happens before any replication is initiated, then brand new Mimivirus particles are produced in the cell cytoplasm accumulating until the eventual lysis of the cell causes their release. Mimivirus first enters the amoeba cell by phagocytosis, meaning that amoeba become infected by way of their own feeding processes. Specifically, the Mimivirus membrane fuses with the cell vacuole membrane and releases what most reasonably is the viral DNA into the cell cytoplasm. Once this DNA enters the nucleus, an amoebic complex forms overtime accumulating mass until that mass begins functioning as the virus factory(VF). This is where DNA encapsidation occurs. The capsid protein L425 is a scaffolding protein that encapsulates the highly accumulated virions in the cytoplasm.[17] This sequence was elucidated through interpretation and comparison of Transmission Electron Microscopy(TEM) slides(Figure 6.) and is still a topic of active research. Through Difference Interference Contrast(DIC) microscopy, researchers were able to topographically visualize 10um tall volcano like features of the virus factory. In figure 7. The DAPI staining in blue corresponds to the presence of DNA. Since the blue regions are highly concentrated to these volcano like structures, it appears that they are a focal point for newly synthesized DNA.
Megavirus Gene Products
All NCLDVs have 5 major defining genes in common. Poxvirus D13 codes for the primary capsid protein, D5 specifies a helicase-primase for the initiation of DNA replication. NCLDVs also share a DNA polymerase elongation sub-unit family protein, an A32 ATPase encoding region and an A2L gene specifying a viral late transcription factor. This suite of genes help to characterize NCLDVs in addition to their large size and a set of 50 other genes that are not common between NCLDVs, but together help to form a suite in a proposed ancestor for all the NCLDVs. These 50 genes also clearly separate NCLDVs phylogenetically from other large viruses which interact pathogenically with Eukaryotic organisms like herpesvirus, in addition to other large prokaryotic and archaea infecting large viruses.[18] NCLDVs also share double-stranded DNA and similar replicative cycles that create viral factories in the cytoplasm.
Megavirus Human Interaction
Although Mimivirus and Megavirus chilensis are known to interact with amoeba and not directly with human hosts, other NCLDVs like the small pox virus have significant human interaction. Small pox is classified as a member of the Poxviridae family, which contains a diverse set of pathogenic viruses that parasitize other animals, such as rabbit myxoma virus and vaccina virus. Poxviridae members also have parasitic interactions with unicellular eukaryotes.[19] Even though Mimivirus doesn’t have a clear pathogenic interaction with humans, unsurprisingly the discovery of Mimivirus was a direct result of research aimed at addressing pneumonia infections associated with the water-tower amoeba that Mimivirus were using as hosts.[20]
Megavirus Chilensis
Megavirus chilensis was recently discovered as pathogenic occupants of Chilean coastal amoeba. It is noteworthy as a distant relative to the NCLDV Mimivirus and currently stands as the largest known megavirus at approximately 700nm in diameter including the icosahedral capsid and virus “hairs”. [21] Even though the structures of Mimivirus and chilensis are remarkably similar, they differ significantly in terms of genetics, with only 50% of residues being conserved on average between any given Mimivirus and chilensis viral particles. In addition to this, chilensis has 594 orthologs with Mimivirus out of the roughly 1000 gene products each NCLDV type makes. This puts chilensis and Mimivirus at the borderline of maximum difference usually considered within a bacterial classification (i.e. firmicutes.). Chilensis is also unique for its three extra amino-acyl tRNA synthetases, relative to Mimivirus. [22]
Virophages
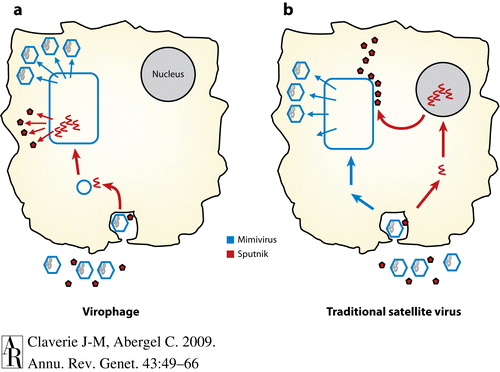
NCLDVs such as Mimiviridae are remarkable discoveries in their own right, but their finding also led to the awareness of the equally remarkable virophages. Virophages are tiny satellite viruses. The first characterized was the sputnik virus, a viral particle only 50nm in diameter and with capsids of icosahedral form. The DNA is similarly small counting in at 18kb, which gives them a small theorized coding capacity of only 21 proteins. A few of these proteins are completely novel in having no homologous nature with other viral genomes. The sputnik virus interacts directly with Mimivirus viral factories inhibiting the NCLDV’s normal replication in amoeba and pathogenicity by 70% and using the viral factories for it’s own reproduction(Fig. 8). Current research hints at the possibility of virophages acting as a part of a NCLDV’s mobile genetic elements, meaning that virophage virions become integrated thoroughly in the genome. This is the case for a different virophage called Lentile virus, which also has transpovirons, that can recombinate with other viruses such as sputnik2. Therefore, NCLDVs create and facilitate the exchange of genes between these virophages, and this interaction could be beneficial in better understanding the complex genomes of NCLDVs.[23]
Conclusion
There is convincing evidence based on phylogenetic analysis of certain target Megavirales genes that researchers use to support the separation of NCLDVs or Megaviruses from the other domains of life. However, contradicting or inconclusive analyses also exist, whether or not these viruses are considered their own new form of life, they serve as a fascinating example for the diversity of life that can occur. In addition, they provide potential knowledge towards the evolution of eukaryotes, and at least have the potential to aid in revolutionizing our understanding of early forms of life.
References
- ↑ http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4083134/pdf/1743-422X-11-120.pdf Abrahão, Jônatas S., Fábio P. Dornas, Lorena Cf Silva, Gabriel M. Almeida, Paulo Vm Boratto, Phillipe Colson, Bernard La Scola, and Erna G. Kroon. "Acanthamoeba Polyphaga Mimivirus and Other Giant Viruses: An Open Field to Outstanding Discoveries." Virology Journal Virol J 11.1 (2014): 120. Web
- ↑ http://www.karger.com/Article/FullText/336562http://www.karger.com/Article/FullText/336562 Colson, Philippe, Xavier De Lamballerie, Ghislain Fournous, and Didier Raoult. "Reclassification of Giant Viruses Composing a Fourth Domain of Life in the New Order Megavirales." Intervirology 55.5 (2012): 321-32. Web. 21 Apr. 2016.
- ↑ http://link.springer.com/article/10.1007%2Fs00705-013-1768-6 Colson, Philippe, Xavier De Lamballerie, Natalya Yutin, Sassan Asgari, Yves Bigot, Dennis K. Bideshi, Xiao-Wen Cheng, Brian A. Federici, James L. Van Etten, Eugene V. Koonin, Bernard La Scola, and Didier Raoult. "“Megavirales”, a Proposed New Order for Eukaryotic Nucleocytoplasmic Large DNA Viruses." Arch Virol Archives of Virology158.12 (2013): 2517-521. Web. 24 Apr. 2016
- ↑ http://virologyj.biomedcentral.com/articles/10.1186/1743-422X-9-161 Yutin, Natalya, and Eugene V. Koonin. "Hidden Evolutionary Complexity of Nucleo-Cytoplasmic Large DNA Viruses of Eukaryotes." Virology Journal Virol J 9.1 (2012): 161. Web. 25 Apr. 2016.
- ↑ http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3291303/pdf/cib-5-102.pdf Legendre, Matthieu, Defne Arslan, Chantal Abergel, and Jean-Michel Claverie. "Genomics of Megavirus and the Elusive Fourth Domain of Life." Communicative & Integrative Biology 5.1 (2012): 102-06. Web. 22 Apr. 2016.
- ↑ http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2263039/pdf/1471-2148-8-12.pdf Moreira, David, and Céline Brochier-Armanet. "Giant Viruses, Giant Chimeras: The Multiple Evolutionary Histories of Mimivirus Genes."BMC Evol Biol BMC Evolutionary Biology 8.1 (2008): 12. Web.
- ↑ http://cid.oxfordjournals.org/content/45/1/95.full.pdf+html Raoult, D., B. L. Scola, and R. Birtles. "The Discovery and Characterization of Mimivirus, the Largest Known Virus and Putative Pneumonia Agent."Clinical Infectious Diseases 45.1 (2007): 95-102. Web. 23 Apr. 2016.
- ↑ http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4083134/pdf/1743-422X-11-120.pdf Abrahão, Jônatas S., Fábio P. Dornas, Lorena Cf Silva, Gabriel M. Almeida, Paulo Vm Boratto, Phillipe Colson, Bernard La Scola, and Erna G. Kroon. "Acanthamoeba Polyphaga Mimivirus and Other Giant Viruses: An Open Field to Outstanding Discoveries." Virology Journal Virol J 11.1 (2014): 120. Web
- ↑ http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3270958/?report=reader Piacente, F., M. Marin, A. Molinaro, C. De Castro, V. Seltzer, A. Salis, G. Damonte, C. Bernardi, J.-M. Claverie, C. Abergel, and M. Tonetti. "Giant DNA Virus Mimivirus Encodes Pathway for Biosynthesis of Unusual Sugar 4-Amino-4,6-dideoxy-D-glucose (Viosamine)." Journal of Biological Chemistry 287.5 (2011): 3009-018. Web. 25 Apr. 2016.
- ↑ http://www.karger.com/Article/FullText/312913 Koonin, Eugene V., and Natalya Yutin. "Origin and Evolution of Eukaryotic Large Nucleo-Cytoplasmic DNA Viruses." Intervirology 53.5 (2010): 284-92. Web. 24 Apr. 2016.
- ↑ http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3291303/pdf/cib-5-102.pdf Legendre, Matthieu, Defne Arslan, Chantal Abergel, and Jean-Michel Claverie. "Genomics of Megavirus and the Elusive Fourth Domain of Life." Communicative & Integrative Biology 5.1 (2012): 102-06. Web. 22 Apr. 2016.
- ↑ http://virologyj.biomedcentral.com/articles/10.1186/1743-422X-9-161 Yutin, Natalya, and Eugene V. Koonin. "Hidden Evolutionary Complexity of Nucleo-Cytoplasmic Large DNA Viruses of Eukaryotes." Virology Journal Virol J 9.1 (2012): 161. Web. 25 Apr. 2016.
- ↑ http://virologyj.biomedcentral.com/articles/10.1186/1743-422X-9-161 Yutin, Natalya, and Eugene V. Koonin. "Hidden Evolutionary Complexity of Nucleo-Cytoplasmic Large DNA Viruses of Eukaryotes." Virology Journal Virol J 9.1 (2012): 161. Web. 25 Apr. 2016.
- ↑ http://link.springer.com/article/10.1007%2Fs00705-013-1768-6 Colson, Philippe, Xavier De Lamballerie, Natalya Yutin, Sassan Asgari, Yves Bigot, Dennis K. Bideshi, Xiao-Wen Cheng, Brian A. Federici, James L. Van Etten, Eugene V. Koonin, Bernard La Scola, and Didier Raoult. "“Megavirales”, a Proposed New Order for Eukaryotic Nucleocytoplasmic Large DNA Viruses." Arch Virol Archives of Virology158.12 (2013): 2517-521. Web. 24 Apr. 2016
- ↑ http://bio.lmu.de/~parsch/evogen/NextGenSeq2008.pdf Mardis, Elaine R. "The Impact of Next-generation Sequencing Technology on Genetics." Trends in Genetics 24.3 (2008): 133-41. Web. 25 Apr. 2016.Mardis, Elaine R. "The Impact of Next-generation Sequencing Technology on Genetics." Trends in Genetics 24.3 (2008): 133-41. Web. 25 Apr. 2016.
- ↑ http://www.sciencedirect.com/science/article/pii/S002220110900086X Claverie, Jean-Michel, Renata Grzela, Audrey Lartigue, Alain Bernadac, Serge Nitsche, Jean Vacelet, Hiroyuki Ogata, and Chantal Abergel. "Mimivirus and Mimiviridae: Giant Viruses with an Increasing Number of Potential Hosts, including Corals and Sponges." Journal of Invertebrate Pathology 101.3 (2009): 172-80. Web. 23 Apr. 2016.
- ↑ http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4083134/pdf/1743-422X-11-120.pdf Abrahão, Jônatas S., Fábio P. Dornas, Lorena Cf Silva, Gabriel M. Almeida, Paulo Vm Boratto, Phillipe Colson, Bernard La Scola, and Erna G. Kroon. "Acanthamoeba Polyphaga Mimivirus and Other Giant Viruses: An Open Field to Outstanding Discoveries." Virology Journal Virol J 11.1 (2014): 120. Web
- ↑ http://link.springer.com/article/10.1007%2Fs00705-013-1768-6 Colson, Philippe, Xavier De Lamballerie, Natalya Yutin, Sassan Asgari, Yves Bigot, Dennis K. Bideshi, Xiao-Wen Cheng, Brian A. Federici, James L. Van Etten, Eugene V. Koonin, Bernard La Scola, and Didier Raoult. "“Megavirales”, a Proposed New Order for Eukaryotic Nucleocytoplasmic Large DNA Viruses." Arch Virol Archives of Virology158.12 (2013): 2517-521. Web. 24 Apr. 2016
- ↑ http://www.karger.com/Article/FullText/312913 Koonin, Eugene V., and Natalya Yutin. "Origin and Evolution of Eukaryotic Large Nucleo-Cytoplasmic DNA Viruses." Intervirology 53.5 (2010): 284-92. Web. 24 Apr. 2016.
- ↑ http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4083134/pdf/1743-422X-11-120.pdf Abrahão, Jônatas S., Fábio P. Dornas, Lorena Cf Silva, Gabriel M. Almeida, Paulo Vm Boratto, Phillipe Colson, Bernard La Scola, and Erna G. Kroon. "Acanthamoeba Polyphaga Mimivirus and Other Giant Viruses: An Open Field to Outstanding Discoveries." Virology Journal Virol J 11.1 (2014): 120. Web
- ↑ http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3198346/pdf/pnas.201110889.pdf Arslan, D., M. Legendre, V. Seltzer, C. Abergel, and J.-M. Claverie. "Distant Mimivirus Relative with a Larger Genome Highlights the Fundamental Features of Megaviridae." Proceedings of the National Academy of Sciences 108.42 (2011): 17486-7491. Web. 24 Apr. 2016.
- ↑ http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3198346/pdf/pnas.201110889.pdf Arslan, D., M. Legendre, V. Seltzer, C. Abergel, and J.-M. Claverie. "Distant Mimivirus Relative with a Larger Genome Highlights the Fundamental Features of Megaviridae." Proceedings of the National Academy of Sciences 108.42 (2011): 17486-7491. Web. 24 Apr. 2016.
- ↑ http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4083134/pdf/1743-422X-11-120.pdf Abrahão, Jônatas S., Fábio P. Dornas, Lorena Cf Silva, Gabriel M. Almeida, Paulo Vm Boratto, Phillipe Colson, Bernard La Scola, and Erna G. Kroon. "Acanthamoeba Polyphaga Mimivirus and Other Giant Viruses: An Open Field to Outstanding Discoveries." Virology Journal Virol J 11.1 (2014): 120. Web
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2016, Kenyon College.