Membrane proteins as mechanisms of immune system evasion in Trypanosoma brucei
Alexandra Stamatoiu
Kenyon College Class of 2014
Introduction

What is Trypanosomiasis?
Human African Trypanosomiasis, commonly referred to as sleeping sickness, is a vector-borne parasitic infection, affecting rural populations in sub-Saharan Africa. The parasite is transmitted through the tse tse fly, which is endemic to this area. African populations (ranging in size from villages to entire regions) that rely heavily on fishing, agriculture, hunting, or other animal husbandry are frequently exposed to the insect and therefore the disease. Trypanosomiasis confers many symptoms, but is most often characterized by drowsiness and an eventual coma that give the disease its name and lead to the death of the host.
Why study it?
Of the many described parasitic human pathogens, the transmitter of Trypanosomiasis has a particularly fascinating relationship to its hosts. As heteroxenous organisms, Trypanosoma parasites require more than one host to successfully complete their life cycles. Specifically, these flagellate protozoa are unicellular and carry out part of their growth inside the gut of the tse tse fly prior to infection of the human bloodstream. The mechanisms through which Trypanosoma species evade the human immune system are complex and not completely defined.
Much research is concerned with the study of the exact modes these unique organisms have for escaping antibody detection and wreaking havoc on their hosts’ health. Perhaps the most important skill Trypanosoma species possess is the ability to manipulate their membrane receptor proteins at a more rapid speed than that with which human immune systems can assemble appropriate antibodies. The complex interactions between membrane surface proteins and the human immune cells responsible for the recognition and eradication of invading organisms are what define trypanosomes as deadly parasites.
The inadequacy of current treatments has spurred on research by associations like the World Health Organization (WHO) aimed at describing this phenomenon in greater detail and at discovering more effective means of understanding the many facets of African sleeping sickness. Novel molecular interactions are presently being discovered that provide further and further insight into trypanosomes and their means of infection.
Various species of the genus Trypanosoma confer disease to humans. The most prevalent and thus most widely studied is Trypanosoma brucei, a eukaryotic flagellate. The parasite is a typical eukaryote in that it contains fully functional mitochondria and other membrane-bound organelles. The mitochondria of the trypanosome include kinetoplasts, disk-shaped networks of condensed circular DNA containing many copies of the mitochondrial genome. Another defined pathogenic species is T. cruzi, which confers what is known as Chagas' disease in humans and animals and is found mostly in North America [5]. Both are human pathogens but the prevalence and rate of infection of T. brucei makes it a more relevant and fascinating research subject.
Disease Origins and Distribution
Two morphologically indistinguishable subspecies of Trypanosoma brucei have been described to cause diverse disease patterns in humans: Trypanosoma brucei rhodesiense (East African sleeping sickness) and T. b. gambiense (West African sleeping sickness).
As demonstrated by Figure 2, the two strains are chiefly found in different regions of Africa and there is currently minimal data in support of overlap in their distribution. Any regions where infringement of territory is indicated are merely speculative.
Predominantly found in regions of eastern and southeastern Africa, T. b. rhodesiense infections are the most prevalent in Tanzania, Uganda, Malawi, and Zambia [11]. Wild animals have been found to be the most effective reservoirs for these trypanosomes, capable of infecting local populations as well as sporadically transmitting to hunters and game park visitors. As a result, approximately one case per year is diagnosed in the United States [11].
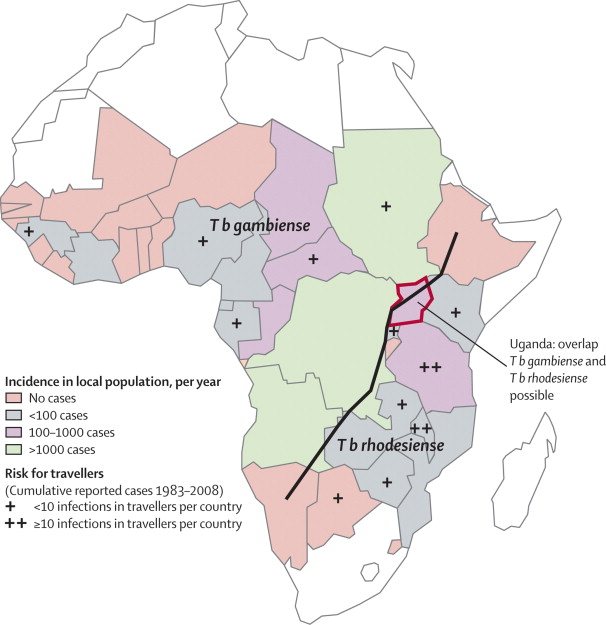
While a few hundred incidences of East African sleeping sickness are reported annually, the majority of sleeping sickness cases in African populations stem from T. b. gambiense, found primarily in central Africa and areas of West Africa [11]. Trypanosomiasis is most common to the poorest rural populations of some of the least developed Central African countries [2]. Of the thousands of cases reported to the WHO each year, more than 95% originate from Democratic Republic of Congo, Angola, Sudan, Central African Republic, Chad and northern Uganda [11].
Mechanism of Infection
The Tse Tse Fly Vector
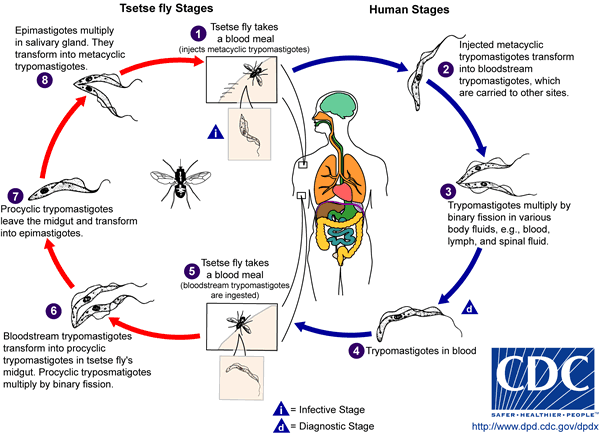
The most common mode of infection is a bite from an infected Glossina palpalis, or tse tse fly (WHO). Tse tse flies occupy mostly rural areas (specifically in woodlands, forests, and vegetation), which is why poorer populations are more exposed to the disease. The fly’s ability to bite and confer the parasite is not inhibited by its sex and it has been reported to only bite during the daylight hours [10]. The parasite uses the fly as an effective way to gain access to the human blood stream. The infected tse tse fly injects the mammalian host skin tissue with metacyclic trypomastigotes that transform into bloodstream trypomastigotes (Fig. 3).
Once inside, T. brucei can multiply freely by binary fission in the blood and lymph. As represented in Figure 3, the trypanosome life cycle in the fly overlaps the parasites infective trajectory in a mammalian host. The tse tse fly ingests bloodstream trypomastigotes from feeding on the blood of infected humans and the parasites transform back into their procyclic forms in the fly midgut. Trypomastigotes multiply and undergo a transformation into epimastigotes, slowly traveling to the fly’s salivary glands where they ready to enter mammalian hosts. This cycle takes around three weeks in the Glossina species (Fig. 4).

While Glossina constitute the vast majority of transmission modes, other mechanisms have been detected. Transmission of the infection from mother to unborn baby has been observed and it could also feasibly be conferred through transfusion of blood or even prolonged blood contact, but such cases are incredibly rare.
Symptoms, Diagnosis, and Treatment
Two subspecies of T. brucei are found to infect humans and each expresses a variation in observable clinical features [6]. Infection by T. brucei corresponds to initial symptoms of headache, weakness, and even joint pain. If it goes untreated, the disease’s intermediate stages accompany cardiovascular problems, kidney disorders, and anemia [11]. In its final phase, Trypanosomiasis can lead to extreme exhaustion and fatigue, coma, and eventually death.
The Center for Disease Control and Prevention (CDC) has described two distinct clinical junctures regarding sleeping sickness. The first is defined by the presence of the parasite solely in the host’s peripheral circulation, before it has fully penetrated the central nervous system (CNS). Once the blood-brain barrier is crossed and the CNS is infected, the disease is said to have entered its second phase. However, because there are two genetically distinct versions of the parasite, there are also two discrete sets of symptoms.
The most fundamental distinction between T. b. rhodesiense (East African sleeping sickness) and T. b. gambiense (West African sleeping sickness) is the rate at which their respective infections proceed. The East African variant of the disease progresses rather rapidly, causing a large chancre (or sore) to develop at the site of parasite entry and causing death within months [11]. T. b. gambiense symptoms appear much more slowly and may be very mild in the beginning, killing the host in no longer than 6-7 years, but more often in about 3 years [11].
African trypanosomiasis is diagnosed largely through laboratory methods. Namely, the diagnosis is straightforward and relies on visualization of the parasite in body fluid through microscopy. T. b. rhodesiense parasites are easily observed in blood, whereas T. b. gambiense is easier viewed from microscopic analysis of lymph node aspirate [11]. Specific drug treatment is directly related to the form of the parasite and the stage of the infection (whether or not the parasite has reached the CNS).
Drugs like pentamidine, suramin, melarsoprol, eflornithine, and nifurtimox have been used in the U. S. to combat African trypanosomiasis [11]. Because instances of relapse have been observed, patients are recommended to have serial examinations of cerebrospinal fluid for up to 2 years post treatment [11]. Sadly, current treatments tend to be inadequate, as drugs intended for late-stage sleeping sickness are highly toxic and no vaccine currently exists that is completely effective in preventing the disease [2].
Protein Trafficking
What is Antigenic Variation?
African trypanosomes modify their surface membranes in order to evade host immune systems, multiplying with every surface change in a process known as antigenic variation [2]. The parasite can even elude acquired immunity during infection because of the rate at which they multiply.
One of the reasons T. brucei is an incredibly fascinating human pathogen is its ability to undergo rapid membrane protein variation and replicate faster than the immune system can build up a defense against it. A healthy immune system can usually generate antibodies in a relatively brief period, but not fast enough to catch the trypanosomes before their protein compositions are altered. In an effort to better quantify this relationship, research has been conducted to analyze and sequence the trypanosome genome from its 11 megabase-sized chromosomes [2]. The genome has been determined to contain about 9068 predicted genes, approximately 900 of which are pseudogenes and close to 1700 are genes specific to T. brucei.
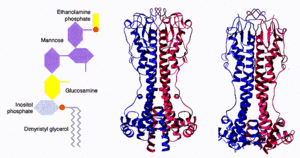
T. brucei surface membranes contain invariant antigens that are shielded by an almost impenetrable coat of 107 copies of a single variant surface glycoprotein (VSG) that the parasite replaces as the immune system accumulates antibodies against it [2]. An illustration of the general structure of trypanosome VSGs is provided in Figure 6.
According to current research, trypanosomes have approximately 806 VSGs, of which most are pseudogenes that the parasites can ectopically recombine to generate expressed mosaic genes. In other words, the dysfunctional genes can be recombined within the trypanosome cell to create functional or coding genes, which allow previously genetically identical cells to express different genotypes. Trypanosomes possess the ability to produce hundreds of different VSG variants, so different in their sequences that host antibodies can’t possibly be effective against all forms [3]. Additionally, every trypanosome contains genes for each possible VSG, thus it does not have to waste time and expend energy with the assemblage of these glycoproteins like the human immune system does during antibody production.
Recent Findings
Current research on the subject of trypanosome membrane protein recycling and its involvement in the pathogen’s ability to successfully infect and avoid immune detection offers a great deal of insight into T. brucei mechanisms of infection. Work recently published by Koumandou et al. (2013) illustrates the importance of intracellular trafficking regarding sleeping sickness infection.
While much material has been described on the subject of T. brucei’s sophisticated VSG gene-switching mechanism, the data from this report implicate invariant surface glycoproteins (ISGs), as well [7]. These surface proteins are much more stable than VSGs, as their name would imply, and are highly relevant for the binding and uptake of drugs aimed at treating trypanosomiasis.
Trypanosome cell surfaces are covered in a high-density coating of VSGs, which inhibits the immune system’s ability to recognize invariant epitopes. Trypanosome gene switching produces VSG proteins that are immunologically distinct from one another and thus the population cannot be entirely eradicated. As such, previously unknown VSG sequences continue to be identified. Specifically, this research produced a set of 29 VSG-related sequences in a tight cluster.
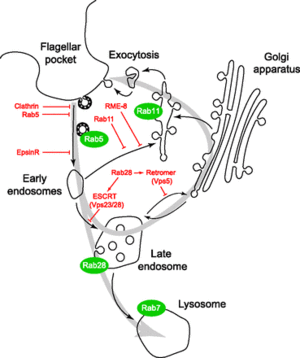
Under normal circumstances, the components of the body’s immune system specialized for antigen recognition (B cells, T cells, and even antibodies) can recognize foreign cells by their antigenic determinant proteins. T. brucei’s thick membranous coat of VSGs helps shield it from immune detection. The antibodies that do manage to bind to the cell surface are quickly cleared in a protein-mediated pathway, specifically referred to as clathrin-mediated endocytosis, or CME (Fig. 7).
Clathrin is heavily implicated in the cellular formation of coated transport vesicles and it has recently been associated with the maintenance of both the VSG coat and the trypanosome’s ability to rid its surface of antibodies. As demonstrated by Figure 7, taken from research by Koumandou et al, Clathrin mediates transfer of antibody-capped VSGs to the cell’s flagellar pocket, where they are endocytosed and further trafficked throughout the endocytic system [7]. The endocytic pathway then sorts the antibodies for degradation and recycles the VSGs back to the plasma membrane, effectively making clathrin a key component of immune system evasion in T. brucei. In addition to these VSG interactions, ISGs have been described to aid in the creation of a therapeutic pathway into the cell (ISGs assist in the uptake of first-line therapeutic agents).
Interestingly, the cell’s invariant surface glycoproteins, counterparts intercalating with VSGs in the membrane, have also been implicated in immune resistance [7]. They exhibit type I transmembrane topology, meaning they anchor to the membrane and their N-terminal ends are targeted to the lumen of the endoplasmic reticulum during synthesis (Fig. 6). They have been found to be receptors involved in the uptake of suramin, a drug compound used as a first-line defense in the treatment of trypanosomiasis.
T. brucei undergoes fairly dramatic membrane protein composition shifts throughout its growth trajectory, utilizing both gene-switching methods and the clearing of antibodies from the cell surface to effectively escape immune detection. The previously described endocytic activity tends to be upregulated when the trypanosome is in the mammalian bloodstream stage compared to when it exists in its insect form. Clathrin is developmentally regulated and its involvement is indicative of its direct relationship to pathogenicity and immune system evasion. As such, Koumandou and colleagues (2013) analyzed additional transcripts in hopes of discovering upregulated genes hinting at further mechanisms of infectivity in mammals. They uncovered an ortholog (or a gene descended from a common ancestral form, differentiating due to a speciation event) of the metazoan RME-8 (“Required for receptor-Mediated Endocytosis 8”), which appears to be significantly overexpressed in parasites in the blood-stage [7]. This ortholog controls some of clathrin’s functions, such as the uncoating of its endosomes and the promotion of retrograde transport of endosomes to the Golgi apparatus.
The function of this cellular component has been described for various model organisms (including Caenorrhabditis elegans, Drosophila melanogaster, Arabidopsis thaliana, and mammalian species) and has been found to perform similarly to what is known about its interaction with trypanosome life cycles (involved mainly in prelysosomal trafficking). Trypanosomes are distantly related to yeast and possess simple, but complete cellular trafficking systems and can thus be studied using similar methods.
Conclusion and Future Research
Trypanosomes in the Context of the Immune System
The field of immunology is a complex one that is constantly expanding and developing. The many components of the immune system perform different functions of varying specificity, of which the most important is arguably the ability to make the distinction between foreign cells and the body's own tissue. This differentiation between extrinsic and intrinsic agents is what makes the immune system equally fascinating and complicated. Human immunity relies heavily on the body's ability to respond to viruses and parasites in order to eradicate pathogens and maintain a consistent level of health. While the innate immune system provides an immediate, albeit non-specific, response to generalized exposure, the adaptive immune system is of greater significance to trypanosomiasis [6]. The acquired immune system is involved in the production of antibodies in response to antigens and is greatly limited by trypanosome protein interactions.
A Pattern of Neglect
Trypanosomiasis is one of the tropical diseases considered to largely be neglected by the rest of the world. Even while the illness is endemic in 36 African countries, organizations like Doctors Without Borders (or, MSF) argue that those affected by the parasite are not receiving the treatment or attention they are due by the international community. A significant number of populations most susceptible to sleeping sickness reside in remote, rural areas and as such cannot readily access health services and treatment.
MSF currently organizes satellite laboratory facilities that specialize in testing rural populations for sleeping sickness. However, due to the relative lack of funding and support dedicated to this disease, the methods that must be used are costly and inefficient. Testing a small village can take hours (in addition to assembling and taking apart the make-shift laboratory required to analyze blood samples) and the current mechanism of testing for trypanosomiasis involves a sample from a lumbar puncture, a particularly painful spine injection.

Apart from the difficulty with merely diagnosing an individual with trypanosomiasis, the medications used to treat the disease are extremely hazardous. For example, until as recently as 2010, the most widely-used drug against African sleeping sickness was melarsoprol, a derivative of arsenic that was reported to have killed 1 in 20 patients. While safer, more effective treatments are currently being developed (like nifurtimox-eflornithine combination therapy, or NECT), there is still a long way to go to fully understand this disease.
With more attention dedicated to the treatment of this widespread infection, methods of treatment and of screening populations could be significantly helped and eventually the prevalence of the disease for these people would decrease.
Hope for the Future
While a vast array of information exists surrounding Trypanosoma brucei membrane protein composition alterations in response to immune detection, novel data can still be assessed pertaining to trypanosome surface proteins. New genes are continually sequenced that provide more and more insight into an area that has the potential to save thousands of human lives. Although medications do currently exist as methods of curing the disease (if caught early enough), trypanosomiasis is a lethal disease that at this point has no definitive means of prevention. Through drug interactions it can be treated but the available medications for late-stage infections are incredibly hazardous and detrimental to the body.
New insight in the study of trypanosome life cycles has been revealed concerning their genome analysis. The C-terminal domains that comprise the VSG proteins are much more degenerate than their N-terminal zones, leading to the creation of hybrid gene formation. This novel description of the process provides further evidence for complex mechanisms of protein coat manipulation. By effectively and accurately describing the communications that make up the minute microbiology of the trypanosome cell, we can essentially reveal vital information about the disease as a whole and about pathogenic parasites, in general.
References
3. Borst, P. "Molecular Basis for Trypanosome Antigenic Variation". Cell. 1982. Volume 29. p. 291-303.
6. Foster J W and Slonczewski J L. Microbiology: An Evolving Science. New York City: W. W. Norton Limited, 2010. Print.
12. Biochemistry of Antigenic Variation in African Trypanosomes.
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2009, Kenyon College.
