Metabolic Processes in Rhodopseudomonas palustris
Introduction
By A. Pearlman
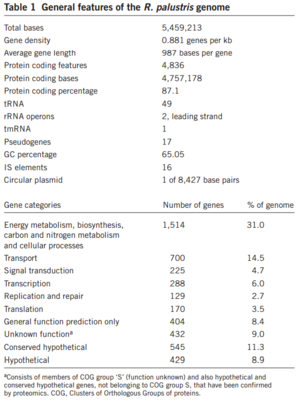
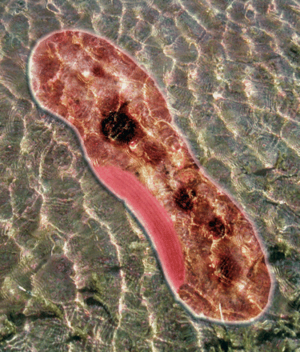
Rhodopseudomonas palustris, recognizable by its purple color, is a gram-negative alpha proteobacterium commonly found in pond and river water as well as the surface of soil and rock (Figure 1). The species is capable of a remarkably wide host of metabolic processes, including aerobic respiration, anxoygenic photosynthesis (Harwood Lab), carbon fixation, nitrogen fixation, and extracellular electron transfer. These modes of metabolism are not all active at once, and are triggered in response to certain environmental cues. In order to support all these processes efficiently, much of its 5.5 million bp genome is devoted to regulation, signaling, and transport (Figure 2). Furthermore, its genome has an unusually smaller number of redundancies (theoretically to maximize the amount of varying information that can be stored) and does not appear to bear the products of many phage genome integrations or horizontal gene transfers, further begging the question of how it was able to evolve so many different metabolisms (Larimer et al. 2004).
R. palustris has two potential reproduction cycles that it may follow, depending on the environmental conditions. Much of the energy used in the reproduction process is derived from light-based metabolism, and is possible in anaerobic and aerobic conditions. In the presence of oxygen, R. palustris will generally undergo budding, resulting in two daughter cells. One of these daughter cells will not be motile and will possess a stalk (derived from the original parent cell), while the other will be motile and is classified as a "swarmer." Under hypoxic conditions, R. palustris cells will develop "intracytoplasmic membranes vesicles" that eventually differentiate into the daughter cells. It should be noted that some degree of light is necessary for this process (Achanta 2010).
The aerobic reproduction proceeds via 4 primary steps:
- First, a bud begins to form opposite of the parent cell's flagellum. This bud is tubular in shape, and extends to up to two times the size of the parent body. The tube was shown to be relatively elastic, and was able to bend around nearby detritus if necessary.
- In the second stage, the far end of the tube-like bud grows to a more spherical shape as its organelles develop more.
- After sufficient growth of the bud, the parent and daughter cells divide in an asymmetric fashion, with the parent cell retaining most of the tube-like appendage. As the division happens, the daughter cell is able to move/swim away from the parent. The parent cell, however, is no longer motile at all.
- In the fourth stage, the daughter cell matures by growing in length to the shape typical of R. palustris and eventually appears identical to the parent cell after enough development, save that it possesses motile ability.
The parent cell still possesses the ability to reproduce normally, and has actually been shown to reproduce faster than before, as it has already developed the tube-like structure where the bud forms (its reproduction time has been shown to be 210 minutes on average). The daughter cells can reproduce as well, but will have to completely develop the budding structure before that is possible (resulting in a reproduction time of about 221 minutes total) (Whittenbury & McLee 1967).
Genomics
Since R. palustris supports such a variety of metabolic systems, much of its genome is accordingly devoted to metabolism. So far, some of the metabolism-related gene regions that allow for oxidation of thiosulfate ions, gaseous elemental hydrogen, carbon monoxide, and carbon dioxide have been found, sequenced, and analyzed. Furthermore, certain regions have been identified that allow for R. palustris to break down a host of organic molecules, such as lignin and a variety of fatty acids. Interestingly, there is code for two separate enzyme systems, each of which appears to confer the same catabolic abilities. As of yet, it is not known why this apparent redundancy exists in a species of microbe that is quite constrained for space in its genome, nor has it been determined if different conditions will trigger one of these systems or the other. Other key features of the R. palustris genome include code for 4 distinct oxidase genes, which are stimulated into expression in aerobic conditions (Larimer et al. 2004).
Species such as R. palustris are quite common in water sanitization facilities, especially in the presence of sunlight, because its metabolism allows it to degrade and utilize much of the waste present in the water. As such, it was hypothesized that a significant portion of the genome would be dedicated to these metabolic processes. The prediction has proved accurate, and the species has been shown to possess the ability to utilize many more classes of organic compounds than were previously imagined (Larimer et al. 2004).
As R. palustris is capable of the Calvin Cycle, enzymes related to this process were, of course, predicted to be coded for in the genome. It turns out that R. palustris is actually the only living organism with two ruBisCo-like proteins, or RLPs, encoded by its genes. Not much has been discovered concerning the purpose of this arrangement, but it is theorized that they are important for metabolism of sulfur compounds (Larimer et al. 2004).
Metabolisms
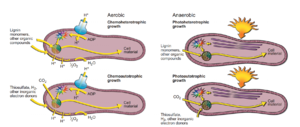
R. palustris can make use of 4 general types of metabolic processes, as shown at right (Figure 3), and they can be categorized as follows:
- Chemoheterotrophy (aerobic conditions)
- Chemoautotrophy (aerobic conditions)
- Photoheterotrophy (anaerobic condtions)
- Photoautotrophy (anaerobic condtions)
(Larimer et al. 2004)
Under chemoheterotropy, an electron transport chain, existing within the cellular membrane, allows for the building of a proton motive force by releasing protons from the cell. ATP synthase enzymes embedded in the cellular membrane drives protons into the cytoplasm, where they are used for regeneration of adenosine triphosphate (ATP) from adenosine diphosphate (ADP). Various organic molecules and compounds (such as lignin) are used as food/electron donors, and are metabolized in a process powered by the previously-generated ATP. Through this metabolic process, the R. palustris cells synthesize building blocks that are later used to make key cellular materials and resources. A byproduct of this process is the formation of water, which is extruded from the cellular membrane (Larimer et al. 2004).
The chemoautotrophic process shares many similarities with the chemoheterotrophic metabolism: there is an electron transport chain within the membrane (which generates the proton motive force), ATP synthase enzymes within the membrane that use protons to re-form ATP from ADP. However, the "food" sources are quite different in this process. In chemoautotrophy, every organic building block that the cell needs originates from carbon dioxide, so this must be absorbed through the cellular membrane into the cytoplasm, where it is eventually used in the central metabolism. The other main differences is the source of electrons: during chemoautotrophic growth, the "food" used is entirely inorganic. R. palustris is capable of using, among other compounds, hydrogen gas and thiosulfate ions as electron donors (the electrons are needed to power the electron transport chain) (Larimer et al. 2004).
Photoheterotrophy in R. palustris (and in general) derives energy from light, but requires more carbon than carbon dioxide alone can provide. As a result, organic compounds similar to those used in chemoheterotrophy are required as a food/electron source (lignin monomers, etc. Also like chemoheterotrophy, ATP provides the immediate energy needed to power the breakdown of this food into building blocks, which in turn are used to generate cell material. However, the main difference is the process used to generate this ATP! Instead of a proton motive force-powered generation being generated by an electron transport chain, the inner cytoplasmic membrane uses pure light as an energy source to pump protons through the membrane and out of the cell. Then, ATP synthase can proceed as usual (Larimer et al. 2004).
Lastly, photoautotrophy involves the fixation of carbon (rather than by oxidizing electron donors in chemotrophy) and is powered by energy obtained from absorbed light (which in turn allows (re)generation of ATP). The process is fairly similar to photoheterotrophy, but uses inorganic sources of electrons and carbon dioxide in its central metabolism instead. For R. palustris, the potential inorganic sources of electrons include hydrogen gas, thiosulfate, and ferrous ions. It is important to note that while the cellular machinery active in this metabolic process can easily obtain electrons from iron, it is more difficult to do so during the chemoautotrophic process (Larimer et al 2004).
Perhaps most interestingly, the photoautotrophic metabolism of R. palustris is fairly unique in that it can actually utilize pure electrical current as an electron "donor" via a specialized enzyme system, a revolutionary process that occurs via extracellular electron transfer (EET). While R. palustris can also obtain electrons from electrical current during the chemoautotrophic metabolism, this is not nearly as common (Bose et. al 2014) -- the EET phenomenon is mostly observed in the presence of light. This is theorized to be the case because the major cellular machinery required for EET are coded for in a regulated region of the genome that is activated by the presence of light (Larimer et al. 2004).
Extracellular Electron Transfer
In general, extracellular electron transfer (EET) involves the use of external electrons obtained from the environment to drive metabolic processes in the absence of oxygen (Girguis), and may involve the movement of electrons both into and out of cells. EET is a particularly nuanced form of obtaining electrons, since they are received from outside of the cell rather than from a "food" source that the cell absorbs, such as sugars. The microbes have developed a system by which electrons are accepted by membrane-bound enzyme systems, which in turn assist in the delivery of electrons to the central metabolic processes of the cell, where cellular materials are constructed. Since this system requires close (if not direct) contact between the microbes and the electron source, bacteria that use EET (including R. palustris) typically do not form **thick** biofilms. In order to use the surface area most efficiently, however, they may form thin biofilms that cover the medium when there is high potential for performing EET (Bose et al. 2014, Hernandez & Newman 2001, & Larimer et al. 2004).
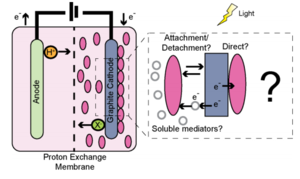
In their natural environment, Rhodopseudomonas palustris has only been observed to use iron as its source for external electrons. However, recent experimentation by Bose, et. al (2014) has shown that R. palustris is very capable of using a variety of other electron-rich compounds as well. The tests involved electrode arrays wired to colonies of the bacteria, which were exposed to surfaces of different compositions (Figure 4). It was discovered that non-ferrous metals and even sulfur compounds were capable of supporting the exact same mode of EET that iron does.
When undergoing metabolism as a result of EET, Rhodopseudomonas palustris generally remain on the surface of the electron source in order to absorb light. However, due to their naturally-conductive properties, they are able to draw electrons up towards them from within the source. The energy derived from light absorption can be used both for powering RuBisCO in carbon fixation (photosyntetically) as well as aiding in the uptake and acquisition of external electrons. However, R. palustris have been shown to obtain electrons from external sources even in the absence of light (albeit not as much).
(Bose et al. 2014)
According to recent research, external electron transfer and its associated metabolic processes are closely linked to cell growth and reproduction, especially in the presence of light. When exposed to electrodes, R. palustris colonies exposed to constant light grew to a density 10 times greater than those in the absence of light over a 5-day period. It is important to note that the light-derived energy was not necessarily used directly in metabolism, but was instead used to drive increased uptake of external electrons; the study determined that the treatment colony did in fact withdraw far more electrons from the substrate than its control counterpart. This result seems to indicate that key cellular processes related to growth and development may rely heavily on the ATP that is formed using light-derived energy (Bose et al. 2014 & Larimer et al. 2004).
However, it was noted that not all of the increased biomass came from EET-related metabolism. It is currently theorized that "assimilation" of carbon dioxide (drawn from the atmosphere) also contributed. Interestingly though, this carbon fixation still occurred more in the light-exposed colonies, meaning it was not simply used as a replacement or alternative when no light is present. Instead, this result suggests that there may be some degree of linkage in the genetic code for carbon fixation and EET processes that is up-regulated by the presence of light. (Bose et al. 2014)
Genetic Basis for EET
The acquisition of external electrons by R. palustris has been closely linked with an operon known as pioABC, which has already been demonstrated as necessary for the process of photoferrotrophy. Three main proteins are coded for and regulated in expression by the pioABC region: PioA, PioB, and PioC. PioA proteins (which are a specific type of cytochrome) can be found in periplasmic space (closer to the outermembrane), PioB exists as a sheet of secondary-structured proteins that cross the outer cellular membrane, and PioC -- an iron-sulfur cluster protein -- exists adjacent to PioA in the periplasmic space (closer to the cytoplasmic membrane). The order of these proteins, moving from the outer membrane inward, is PioB, PioA, PioC -- this is the path that electrons follow into the cell during uptake from the environment (Bose et al. 2014).
The pioABC system was shown to be directly associated with approximately 30% of external electron transfer in the previously-described study (Figure 5), as strains with a defective pioABC system obtained about 7/10s of the electrons obtained by the wild-type strain. Furthermore, it appears that each of the three Pio proteins have different levels of expression depending on what other metabolic processes are occuring, also shown in the figure at right. It is important to note that, even though pioABC does not account for the majority of electron uptake, it is likely provides more ability for uptake than any other one system or operon. Additionally, while the aforementioned pioABC experiment only examined R. palustris, there are genetically similar regions in a host of other bacterial species, suggesting that the ability to undergo external electron transfer may not be confined to close relatives R. palustris (Bose et al. 2014).
It is important to note that while most recent research regarding EET was performed using R. palustris, there is compelling evidence that other species of microbes are at the very least capable of similar processes, and may in fact use them commonly as well. This has been determined via comparative genomic analysis, which has identified genetic regions similar to the PioABC operon in the likes of other alpha proteobacteria, such as the following:
- Rhodomicrobium vannielii
- Azospirillum amazonense
- Magnetospirillum magnetotacticum
Similar findings exist for several species of bacteria in classes beta proteobacteria, gamma proteobacteria, and delta proteobacteria. Furthermore, the results indicate that even bacteria in other phyla may possess operons similar in function to PioABC, including:
- Acidobacteria
- Solibacter usitatus
- Terriglobus saanensis
(Bose et. al 2014, Supplementary Data)
In addition to EET being regulated by other genes, it is logical to assume that changes in expression of certain proteins might be triggered by EET itself. As it turns out, there is compelling evidence that the physical occurrence of external electron uptake actually stimulates/up-regulates other regions of the genome. Using WT-illumination, the Bose et. al team discovered several specific genetic regions that were much different when the R. palustris were engaging in EET. For example, a protein similar to pioC was found to undergo an increase in expression in treatments exposed to light. This seems to suggest that, under conditions that allow for a large growth rate, the PioABC system may not be "enough" to support the colony development alone. As a result, homologous systems may be triggered in the event of a very readily available electron source in order to make efficient use of the resource.
Interestingly, several other enzymes that were not directly associated with EET appear to have stimulated expression while it occurs. One such enzyme is ruBisCo (specifically form I). The function of this specific enzyme is to catalyze the reaction between carbon dioxide and RuBP to yield 2 molecules of 3PG as part of the Calvin Cycle, which ultimately yields G3P through a reduction process involving NADPH. It has been posited that the increased expression of ruBisCo is made possible by the increased presence of ATP, which in turn is 1) possible due to the presence of light and 2) necessary to utilize the plethora of electrons being obtained. Since G3P is associated with a portion of the Calvin Cycle that involves reduction, the researchers predict that the upregulation of ruBisCo (and consequent additional G3P production) assists in keeping the redox system within the cells balanced in the event of significant electron uptake (Bose et al. 2014).
It should be noted that while EET does indeed stimulate ruBisCo form I expression, this only occurs in the presence of light; in the dark, expression does not change with EET -- this is largely unexplored, but may indicate that only a subset of EET-related genes have been analyzed in this species as of yet (Bose et al. 2014).
References
- Achanta, S. (2010). Rhodopseudomonas palustris.
- Bose, A., Gardel, E., Vidoudez, C., Parra, E., & Girguis, P. (2014). Electron uptake by iron-oxidizing phototrophic bacteria. Nature Communications, 5
- Harwood, C. (2011). Harwood Lab: Rhodopseudomonas palustris
- Hernandez, M., & Newman, D. (2001). Extracellular electron transfer. Cellular and Molecular Life Sciences CMLS, 58(11), 1562-1571.
- Hunter, C. N. (2008). The purple phototrophic bacteria. Springer.
- Larimer, F. W., Chain, P., Hauser, L., Lamerdin, J., Malfatti, S., Do, L., Lang, A. S. (2004). Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nature Biotechnology, 22(1), 55-61.
- Oda, Y., Star, B., Huisman, L. A., Gottschal, J. C., & Forney, L. J. (2003). Biogeography of the purple nonsulfur bacterium Rhodopseudomonas palustris. Applied and Environmental Microbiology, 69(9), 5186-5191.
- Reguera, G., McCarthy, K. D., Mehta, T., Nicoll, J. S., Tuominen, M. T., & Lovley, D. R. (2005). Extracellular electron transfer via microbial nanowires. Nature, 435(7045), 1098-1101.
- Yang, Y., Xu, M., Guo, J., & Sun, G. (2012). Bacterial extracellular electron transfer in bioelectrochemical systems. Process Biochemistry, 47(12), 1707-1714.