Methanosarcina
A Microbial Biorealm page on the genus Methanosarcina
Methanogens overview
Classification
Higher order taxa:
Archaea; Euryarchaeota; Euryarchaeota orders incertae sedis; Methanosarcinales; Methanosarcinaceae
Species:
Methanosarcina acetivorans, M. baltica, M. barkeri, M. lacustris, M. mazei, M. semesiae, M. siciliae, M. thermophila, M. vacuolata
|
NCBI: Taxonomy Genome: M. acetivorans M. barkeri M. mazei |
Description and Significance
Methanosarcina spp. are anaerobic methanogens that can form multicellular colonies. They can be found in a multitude of environments including the rumen in cows, sheep, goats, deer, and the large intestine in humans. Recently, there has been a study on M. barkeri, because evidence of a 22nd amino acid named pyrrolysine has been detected. This protein was located in the active site of the enzyme methogenic methylamine methyltransferase, which catabolizes methylamines leading to methane production.
Genome Structure
Clearly containing the largest archaeal genome (4th largest of Prokaryotes) with 5,751,492 bp, M. acetivorans uses these genes for a multitude of different properties not shown by other archaeons. Another sequenced species, M. 'mazei also has a large genome in relation to other archaeons with 4,096,345 bp. An interesting discovery in the M. acetivorans genome was the presence of chaperonins GroEL/GroES that were believed to occur only in bacteria and eukaryotic cell organelles of bacterial ancestry. Three other main chaperoning systems were discovered as well. A pivotal question is then did M. acetivorans receive the genes by inheritance or by lateral transfer from a bacterium, which was suggested for M. mazei. The answer could dictate whether the common ancestor had these genes or whether they were a bacterial product that have been transferred.
Cell Structure and Metabolism
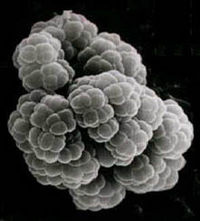

All the other methanogens can utilize no more than two methanogenic substrates and possess a single pathway for methanogenesis. Methanosarcina, on the other hand, has all three known pathways for methanogenesis and can utilize no less than nine methanogenic substrates. M. barkeri and M. mazei are autotrophic, but M. acetivorans is not.
It also has a number of distinct morphological forms including single cells with and without a cell envelope, as well as multicellular packets and lamina. The packets and lamina showed internal morphological diversity, indicating possible cell differentiation. The fact that cells in the lamina secrete different extracellular material gives light to possible cell specialization as well. They are coccoid and have cell walls of protein, often having an external wall of a heteropolysaccharide. Most Methanosarcina spp. are surrounded by a polymeric network of methanochondroitin that is external to an S-layer. The term "matrix" has been proposed to describe this structure. It has been demonstrated by Xun et al. (1990, as cited in Ferry p.89) that the methanochondroitin causes cell-cell adhesion.
Ecology
Methanosarcina display environmental diversity. It has been located in freshwater and marine sediments, decaying leaves and garden soils, oil wells, sewage and animal waste digesters and lagoons, thermophilic digesters, feces of herbivorous animals, and the rumens of ungulates. Further, the species of Methanosarcina grow between 1 to 35°C with the optimal temperature of around 25°C.
References
Galagan et al. 2002. The Genome of M. acetivorans Reveals Extensive Metabolic and Physiological Diversity. Genome Research 12: 532-542.
Conway de Macario, E., D. L. Maeder, and A.J.L. Macario. 2003. Breaking the mould: archaea with all four chaperoning systems. Biochemical and Biophysical Research Communications 301: 811-812.
Ferry, James editor. Methanogenesis: Ecology, Physiology, Biochemistry, & Genetics. Chapman & Hall Inc, New York, 1993.
Atkins, J.F. and R. Gesteland. 2002. The 22nd Amino Acid. Science vol 296: 1409-1410.
Cavicchioli, R. "Cold-adapted archaea" Nature Reviews Microbiology. May, 2006, Volume 4. p. 331-343.
