Microbewiki:Deinococcus radiophilus
- Deinoccocus radiophilus
Introduction
In order to survive, humans require mild environments like neutral pHs, (pH 7) low salinity, (close to 0% salt) or warm temperatures. (37°C) This is similar to many single cell life forms, but not all. Some microbes that are known as extremophiles have evolved to tolerate unhospitable environments like extreme pHs, (pH 2/pH 10) salt concentrations, (5% salt concentration) and temperatures. (60°C+) Although many of these extremophiles have been heavily researched and annotated, some scientists have found microbes in unusual extreme conditions. Examples of these conditions are high UV, radiation, and oxidative environments. These conditions can be harsher than typical extreme conditions, yet certain organisms can flourish. An example of this would be Deinococcus radiophilus, although we have not found a common habitat the microbe resides in, we know that it can be resistant to UV, radiation, and high salt concentration. Thus, making it likely to outcompete other microbes under these conditions.
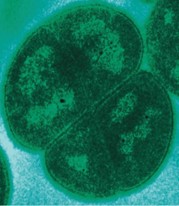
There is no publicly available image of Deinococcus radiophilus but it shares many morphological features with Deinococcus radiodurans, as seen to the left in a transmission electron micrograph. D. radiophilus is an orange-red, gram-positive, tetrad cocci.
Classification
Higher Order Taxa: Bacteria; Deinococcota; Deinococci; Deinococcales; Deinococcaceae.
Species: Deinococcus raidophilus
NCBI Genome: [Genome]
JGI Genome: [Genome]
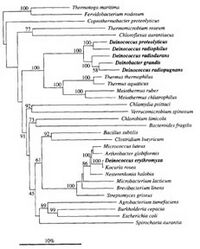
Phylogenetic tree of Deinococcus: Phylogenetic diversity of the Deinococci as they extracted DNA, amplified 16S rDNA with PCR, and sequenced the PCR products. From this analysis, Deinococcus proteolyticus has been found to be the closest relative to D. radiophilus.(2) While 16S RNA sequencing can be a useful phylogenetic method; it could miss key traits that differentiate Deinococci from other microbe, like their multiple stress response genes.

Ecological Habitat
The habitat of the mesophilic members of the Deinococci family, including D. radiophilus, has not been defined.(3) Although the exact habitat of mesophilic Deinococci is unknown, researchers looking at Deinoccocus radiodurans, have noted that the radio-resistances of species withing the family probably evolved independently from each other.4 Additionally, the researchers showed multiple Deinococcaceae species isolated from deserts, high UV environments.(4)
Significance to the Environment
D. radiophilus was isolated from an irradiated Bombay Duck (Harpadon nehereus). A fish found in Indo-Pasific waters.6 Multiple papers have found multiple bacterial niches within the Bombay Duck.(7,8) In particular, in the 1953 study, multiple species of Microccocus were isolated from dried meat. (8) This is of note since Deinococci were originally identified as Micrococci, with the first member isolated 3 years later in 1956. (9)
While the ecological habitat of D. radiophilus remains unknown, it has been isolated from irradiated Bombay Duck (Harpadon nehereus) fillets. (5) If D. radiophilus commonly passes through Harpadon nehereus transiently, the common methods of drying the meat in the sun by humans would select for better desiccation and UV resistances. Deinococcus radiophilus has been isolated from an irradiated Bombay Duck (Harpadon nehereus), which are found in India.
Relation to Bacteriophages
Deinococcus radiophilus has shown to code for an incomplete genome of a prophage named PHAGE_Lactob_Ldl1_NC_026609(2) In addition, Deinococcus radiophilus codes for three restriction enzymes; DraI, DraII, and DraIII. Enzymes used by bacteria to destroy foreign DNA. This supports that Deinococcus radiophilus could have a relationship with bacteriophages and has found strategies to avoid being parasitized to survive and thrive in its niche. This could be relevant since prophages are a sign of temperate phages, which are bacteriophages that insert their genomes into their host. Waiting to undergo lytic reproduction, using their bacterial host as a phage factory, killing the host.
Cell Structure
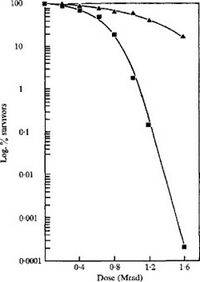
Deinococcus radiophilus is Gram positive, (10,11) and forms tetrads that divide into two plains. They are 1-2µm in diameter and are nonmotile. (10) They exhibit smooth, convex, and regular edged colonies. Additionally, the colonies are orange-red color, plausibly due to the carotenoids it exhibits. These molecules have been known to absorb UV, (12) possibly contributing to Deinococcus radiophilus' UV resistance. In addition to UV, Deinococcus radiophilus is also resistant to gamma radiation, able to tolerate 1.5Mrads.11
Genome Structure, Content, and Gene Expression
Deinococcus radiophilus genome consists of a single chromosome and six plasmids, and it is 2.79Mb long. (13) The genome is circular, and its GC content is 62.59%. D. radiophilus can tolerate NaCl (table salt) and is resistant to up to 1.5 Mrads of gamma radiation. (10) It was sequenced at Hubbard Center for Genome Studies, Durham, NH, USA. This group did not specify why they sequenced the genome. But another group of researchers sequenced the genome to look for prophage related plasmids, but their paper remains unpublished. Analyzing Deinococcus radiophilus' genome we see that unlike other Deinococci, D. radiophilus lacks the reduction of nitrate, and cannot produce acid from glucose in standard media. It has remarkable DNA repair abilities, it also codes for useful restriction enzymes. (11)
Significance to Humans
Deinococcus radiophilus and other members in the Deinococcus family has shown us how single celled organisms can thrive in radioactive conditions. In particular, a few studies have looked into the UV resistance of D. radiophilus. Additionally, D. radiophilus has future possible roles in bioremediation of radioactive materials and sites. As well as possibly transforming D. radiophilus' radiation/UV/oxidative stress resistance machinery into other organisms.
Due to its remarkable DNA repair abilities and/or other qualities like: Resistances to UV, ionizing radiation, and oxidative stress;14 Deinococcus radiophilus is resistant to radioactive materials, thus it would be used for bioremediation purposes. Additionally, by better understanding their radioactive resistance mechanisms, we might be able to incorporate them in organisms of interest.
Interesting Feature: As stated before, the most interesting feature of D. radiophilus is its many resistances. In particular, previous experimentation has shown D. radiophilus having a better resistance to gamma radiation than D. radiodurans, the posterchild of radiation resistance. (5) Additionally, D. radiophilus exhibits a tolerance to media supplemented with 5% NaCl, while D. radiodurans can only survive up to 1% NaCl. (5) This suggests that D. radiophilus could be used over D. radiodurans in certain bioremediations environments like high salinity, or higher gamma radiation. Future research might want research D. radiophilus tolerance to other types of radiation. Additionally, research into transplanting D. radiophilus resistance machinery into other organisms might be useful.
Edited by Alejandro X. Spittel, a @MicrobialTowson student of Dr. Anne M. Estes at Towson University. Template adapted from templates by Angela Kent, University of Illinois at Urbana-Champaign and James W. Brown, Microbiology, NC State University.
- References
- Daly M. Deinococcus radiodurans [Internet]. Wikipedia. 2007. Available from: https://upload.wikimedia.org/wikipedia/commons/7/73/Deinococcus_radiodurans.jpg.
- Rainey FA, Nobre MF, Schumann P, Stackebrandt E, Da Costa MS. Phylogenetic Diversity of the Deinococci as Determined by 16S Ribosomal DNA Sequence Comparison. International Journal of Systematic Bacteriology [Internet]. 1997 Jan 1;47(2):510–4. Available from: https://www.microbiologyresearch.org/content/journal/ijsem/10.1099/00207713-47-2-510
- Battista JR, Rainey FA. Deinococcus. In: Bergey's Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Inc.; 2015. p. 1–13.
- Blasius M, Hübscher U, Sommer S. Deinococcus radiodurans: What Belongs to the Survival Kit? Critical Reviews in Biochemistry and Molecular Biology [Internet]. 2008 Oct 11;43(3). Available from: https://www.tandfonline.com/doi/full/10.1080/10409230802122274
- Lewis NF. Studies on a Radio-resistant Coccus Isolated from Bombay Duck (Harpodon nehereus). Journal of General Microbiology. 1971 Apr 1;66(1):29–35.
- FishBase. Harpadon nehereus (Hamilton, 1822) Bombay-duck [Internet]. FishBase. [cited 2023 May 4]. Available from: https://www.fishbase.se/summary/SpeciesSummary.php?ID=260&AT=Bombay%2BDuck
- Hossain TJ, Chowdhury SI, Mozumder HA, Chowdhury MN, Ali, Ferdausi, et al. Hydrolytic Exoenzymes Produced by Bacteria Isolated and Identified From the Gastrointestinal Tract of Bombay Duck. Frontiers in Microbiology [Internet]. 2023 Aug 26;11. Available from: https://www.frontiersin.org/articles/10.3389/fmicb.2020.02097/full
- Bhat JV, Albuquerque MJ. Proceedings / Indian Academy of Science. 1953;38(3):101–8.
- Gerber E, Bernard R, Castang S, Chabot N, Coze F, Dreux-Zigha A, et al. Hydrolytic Exoenzymes Produced by Bacteria Isolated and Identified From the Gastrointestinal Tract of Bombay Duck. Journal of Applied Microbiology [Internet]. 2015 Jul 1;119(1):1–10. Available from: https://academic.oup.com/jambio/article/119/1/1/6717070
- Brooks BW, Murray RG. Nomenclature for “Micrococcus radiodurans” and Other Radiation-Resistant Cocci: Deinococcaceae fam. nov. and Deinococcus gen. nov., Including Five Species. International Journal of Systematic Bacteriology. 1981 Jul 1;31(3):353–60.
- Maynard CR, MacLea KS. Genome Sequence of the Radiation-Resistant Bacterium Deinococcus radiophilus ATCC 27603T. Microbiology Resource Announcements [Internet]. 2019 Jul 25;8(30). Available from: https://journals.asm.org/doi/10.1128/MRA.00627-19
- Reis-Mansur MC, Cardoso-Rurr JS, Silva JV, De Souza GR, Cardoso VD, Schultz J, et al. Carotenoids from UV-resistant Antarctic Microbacterium sp. LEMMJ01. Scientific Reports [Internet]. 07/02/ 2019;9(1). Available from: https://www.nature.com/articles/s41598-019-45840-6
- Binomica Labs. Deinococcus Radiophilus [Internet]. National Center for Biotechnology Information. [cited 2023 Apr 9]. Available from: https://www.ncbi.nlm.nih.gov/genome/?term=Deinococcus+Radiophilus
- Yun YS, Lee YN. Purification and some properties of superoxide dismutase from Deinococcus radiophilus, the UV-resistant bacterium. Extremophiles: life under extreme conditions. 2004 Apr 23;8(3):237–42.