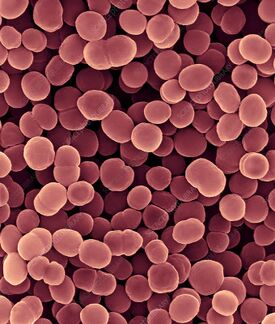
Moraxella catharralis
Classification
Bacteria; Pseudomonadota; Gammaproteobacteria; Pseudomonadales; Moraxellaceae.
Species
Moraxella catarrhalis
Description and Significance
The gram-negative, aerobic diplococcus Moraxella (Branhamella) catarrhalis, formerly known as Neisseria catarrhalis or Micrococcus catarrhalis, is frequently encountered as a pathogen of the upper respiratory tract (Verduin et al., 2002). Since its discovery in 1896, the bacterium has developed into a true pathogen and is now recognized as a significant contributor of upper respiratory tract infections in some of the more healthy young and senior citizens (Constantinescu, 2021).
M. catarrhalis is also a significant contributor to lower respiratory tract infections, especially in individuals with chronic obstructive pulmonary disease (COPD). It is a major contributor to acute exacerbations of chronic obstructive pulmonary disease (COPD), acute bacterial rhinosinusitis, and strep throat in children (Murphy et al., 2005).
Genome Structure
1,863,286 nucleotides make up the 1,886 protein-coding genes in the RH4 genome. The genomes of the ATCC 43617 and RH4 strains are comparable, demonstrating the high degree of gene conservation between them. The 16S rRNA and multilocus sequence typing in silico phylogenetic analyses demonstrate that RH4 is a member of the phylogenetic lineage of the Moraxella catarrhalis.
Since the genes encoding the enzymes needed for various pathways, cycles, and acetate metabolism are present, RH4 depends on fatty acid and acetate metabolism. The routes for acquiring iron, nitrogen metabolism, and oxidative stress responses are some of the pathways crucial for survival under difficult conditions. The ATCC 43617 strain is categorized as a 16S rRNA type 1 strain, just like RH4 and RH4's genome has a 41.7% GC content (Vries et al., 2010).
Cell Structure, Metabolism and Life Cycle
The inferred metabolism of Moraxella catarrhalis has an uncommon characteristic in that it appears incapable of using any exogenous carbohydrates. The absence of entire glycolytic pathways, all carbohydrate catabolic enzymes, and all carbohydrate transport systems in this bacterium shows Moraxella catarrhalis does not consume any of the carbohydrate types and does not create acid from glucose. Gluconeogenesis is necessary since exogenous carbohydrates cannot be used. The entire set of enzymes needed for aerobic metabolism is present in Moraxella catarrhalis. The ATP synthase, glyoxylate cycle, electron transport system, and citric acid cycle are all present and functional in this bacteria.
The way M. catarrhalis uses acetate is associated with a working glyoxylate cycle. In an environment with low-oxygen or a microaerophilic environment nitrate reductase is present, which is compatible with the reported use of nitrate by M. catarrhalis. There are some peculiar features of nitrogen metabolism, such as the fact that M. catarrhalis appears to contain both glutamate dehydrogenase and glutamate synthase-glutamine synthase routes for ammonia absorption. M. catarrhalis has the genes for these enzymes, although it is unclear if it can assimilate ammonia. It lacks all regulators linked to nitrogen limitation in organisms like Escherichia coli. High levels of ammonia cannot be assimilated by M. catarrhalis. Although the genes for ammonia assimilation enzymes are present, it is unclear whether they actually work. They might only be used to synthesize glutamate and glutamine, and they might not be sufficiently expressed to take part in the digestion of ammonia. All amino acids, with the exception of proline and arginine, are synthesized by M. catarrhalis through complete pathways (Wang., 2007).
Ecology and Environment
M. catarrhalis relies on acetate, lactate, and fatty acids for growth, and it is considered an arginine auxotroph. Adults with illnesses or an autoimmune disease typically carry M. catarrhalis in their respiratory passages (Simmons et al., 2012). It binds to the host cell through a trimeric autotransported adhesion and is non-motile.
15%–20% of acute otitis otitis media outbreaks are brought on by the human pathogen Moraxella catarrhalis, which is also a major cause of otitis media in newborns and young children. In the US, M. catarrhalis is thought to be the source of 2-4 million adult cases of chronic obstructive pulmonary disease. M. catarrhalis may go unnoticed in samples from the human respiratory system because it mimic Neisseria species in culture. Infancy and childhood are when upper respiratory tract colonization is most common, while adulthood is when it declines significantly (Murphy et al., 2009).
References
De Vries, S. P., van Hijum, S. A., Schueler, W., Riesbeck, K., Hays, J. P., Hermans, P. W., & Bootsma, H. J. (2010). Genome analysis of moraxella catarrhalis strain RH4, a human respiratory tract pathogen. Journal of Bacteriology, 192(14), 3574–3583. https://doi.org/10.1128/jb.00121-10
Michael Constantinescu, M. D. (2021, August 16). Moraxella catarrhalis infection. Background, Pathophysiology and Etiology, Epidemiology. Retrieved November 16, 2022, from https://emedicine.medscape.com/article/222320-overview#:~:text=Moraxella%20catarrhalis%20is%20a%20gram,Branhamella%20of%20the%20genus%20Moraxella.
Murphy, T. F., Brauer, A. L., Grant, B. J., & Sethi, S. (2005). moraxella catarrhalis in chronic obstructive pulmonary disease. American Journal of Respiratory and Critical Care Medicine, 172(2), 195–199. https://doi.org/10.1164/rccm.200412-1747oc
Murphy, T. F., & Parameswaran, G. I. (2009). moraxella catarrhalis,a human respiratory tract pathogen. Clinical Infectious Diseases, 49(1), 124–131. https://doi.org/10.1086/599375
Simmons, J., & Gibson, S. (2012). Bacterial and mycotic diseases of nonhuman primates. Nonhuman Primates in Biomedical Research, 105–172. https://doi.org/10.1016/b978-0-12-381366-4.00002-x
Verduin, C. M., Hol, C., Fleer André, van Dijk, H., & van Belkum, A. (2002). moraxella catarrhalis : From emerging to established pathogen. Clinical Microbiology Reviews, 15(1), 125–144. https://doi.org/10.1128/cmr.15.1.125-144.2002
Wang, W., Reitzer, L., Rasko, D. A., Pearson, M. M., Blick, R. J., Laurence, C., & Hansen, E. J. (2007). Metabolic analysis of moraxella catarrhalis and the effect of selected in vitro growth conditions on global gene expression. Infection and Immunity, 75(10), 4959–4971. https://doi.org/10.1128/iai.00073-07
Author
Page authored by Student Savannah Brittain at UNC Wilmington.