Mycoplasma fermentans
Template in wiki format under LRMoore Prokaryote Template:
A Microbial Biorealm page on the genus Mycoplasma fermentans
Classification

Higher order taxa
Domain (Bacteria), Phylum (Tenericutes), Class (Mollicultes), Order (Myclosplasmales), Family (Mycoplasmatacea), Genus (Mycoplasma), Species (fermentans) NCBI
Species
Mycoplasma ferementans
Description and significance
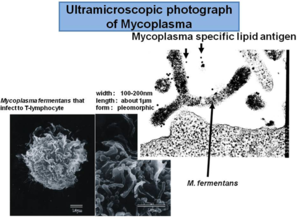
All mycoplasmas M. ferementans are wall-less Gram positive bacteria that colonize human mucus surfaces [1]. M. ferementans are morphologically distinguished from other bacteria by their small size (0.3-0.8 um in diameter) and lack of cell wall [2] (figure 1) . It is mostly found in the genitourinary and respiratory tracts [3] . It is also known for causing arthritis problem in humans [3]. They are surrounded by a lipoprotein membrane which is the responsible for causing inflammatory reactions [4]. These are pathogens that affect humans and the mechanism of infection is not well known yet. It is also believed that this microbe enhances human immunedeficient virus (HIV) by inducing the viral infection [5]. .
Genome structure
Four strains of M. ferementans M64, JER and PG18, P140 have been isolated, characterized, and studied. Based on National Center of Biotechnology Information (NCBI), the entire genome of M. ferementans M64 has been fully sequenced and it is known to have 1,118,751 base pairs that encode for 1050 protein genes, the entire genome is circular [6]. The genome structure of M. ferementans has been compare with other strain called M. fermentans strain JER (977,524 bp) which is smaller. It has a low 26.95% GC content , which is important to distiguish mycoplasma among other genus [7].
Metabolism
Different strains of M. ferementans have been isolated and studied to determine the biochemical variability of this microbe [8]. It is known that this microbe is facultative anaerobe (no oxygen is required for growth but it can grow in the presence of oxygen). There is a lot of variability in the substrate used for metabolic processes. Most of the M. ferementans strains studied until now are able to use glucose during metabolic pathway, but they are also capable of using fructose or arginine, which varies among strains [8].
Ecology
M. ferementans have been isolated from Synovial Fluids of patient with arthritis and also from patients HIV positive, urinary tract and respiratory tract [3, 4, 6]. It have been culture using SP4 media containing pig serum (inactivated) and a pH of 7.6 [8]. The growth condition is around 37ᵒC, which is the body temperature of its host (humans).
Pathology
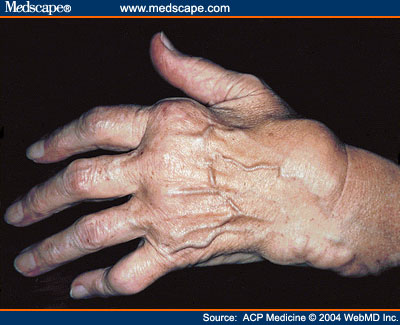
M. ferementans is a pathogenic microbe that affects humans and is usually found in the genital area, and necrotizing tissue [9]. M. ferementans are being linked to diseases like rheumatoid arthritis (figure 2). Also studies suggest that M. ferementans causes chronic fatigue in patient [3]. Some of the strains studied like M. ferementans P.140 are known for causing severe respiratory infections and it has been demonstrated that the infection can travel through the blood to other organs such as the kidney, heart, and the brain [9]. It was discovered that during infection M. ferementans attack cells on the immune system (figure3), such as B cell, causing inflammatory reactions [4]. In humans it is being investigated that M. ferementans is able to enhance HIV replication, through the lipid-associated proteins present on their membrane [5]. M. ferementans seems to enhance the activation of Long term repeat gene, which is important during HIV infection, thorough a protein called toll receptor [5].
References
[1] Marianne in Chinghingyong, Christopher V. Hughes, Detection of Mycoplasma fermentans in human saliva with a polymerase chain reaction-based assay, Archives of Oral Biology, Volume 41, Issue 3, March 1996, Pages 311-314, ISSN 0003-9969, 10.1016/0003-9969(96)84556-0. (http://www.sciencedirect.com/science/article/pii/0003996996845560)
[2] Stothard P, Van Domselaar G, Shrivastava S, Guo A, O'Neill B, Cruz J, Ellison M, Wishart DS (2005) BacMap: an interactive picture atlas of annotated bacterial genomes. Nucleic Acids Res 33:D317-D320
[3] Vojdani, A., Choppa, P.., Tagle, C., Andrin, R., Samimi, B., and Lapp, C.. (1998). Detection of Mycoplasma genus and Mycoplasma fermentans by PCR in patients with Chronic Fatigue Syndrome. Fems Immunol. Med. Microbiol. 22, 355–365.
[4] Gerlic, M., Horowitz, J., Farkash, S., and Horowitz, S. (2007). The inhibitory effect of Mycoplasma fermentans on tumour necrosis factor (TNF)-alpha-induced apoptosis resides in the membrane lipoproteins. Cell. Microbiol. 9, 142–153.
[5] Shimizu, T., Kida, Y., and Kuwano, K. (2004). Lipid-associated membrane proteins of Mycoplasma fermentans and M. penetrans activate human immunodeficiency virus long-terminal repeats through Toll-like receptors. Immunology 113, 121–129.
[6] Shu, H.-W., Liu, T.-T., Chan, H.-I., Liu, Y.-M., Wu, K.-M., Shu, H.-Y., Tsai, S.-F., Hsiao, K.-J., Hu, W.S., and Ng, W.V. (2011). Genome Sequence of the Repetitive-Sequence-Rich Mycoplasma fermentans Strain M64▿. J. Bacteriol. 193, 4302–4303.
[7]Rechnitzer, H., Brzuszkiewicz, E., Strittmatter, A., Liesegang, H., Lysnyansky, I., Daniel, R., Gottschalk, G., and Rottem, S. (2011). Genomic features and insights into the biology of Mycoplasma fermentans. Microbiol. Read. Engl. 157, 760–773.
[8] Afshar, B., Nicholas, R. a. j., Pitcher, D., Fielder, M. d., and Miles, R. j. (2009). Biochemical and genetic variation in Mycoplasma fermentans strains from cell line, human and animal sources. J. Appl. Microbiol. 107, 498–505.
[9] Yáñez, A., Martínez-Ramos, A., Calixto, T., González-Matus, F.J., Rivera-Tapia, J.A., Giono, S., Gil, C., and Cedillo, L. (2013). Animal model of Mycoplasma fermentans respiratory infection. Bmc Res. Notes 6, 9.
Edited by Catherine Lobo of Dr. Lisa R. Moore, University of Southern Maine, Department of Biological Sciences, http://www.usm.maine.edu/bio
