Nanotubes facilitate intercellular signaling in eukaryotic and prokaryotic cells
Overview
<http://www.sciencedirect.com/science/article/pii/S009286741100016X>.
By Daniel Jurgens
Cells encourage diversity by using several different molecular signaling mechanisms to transfer information, nutrients, organelles, or parasites. Many of these processes require direct cellular interaction for signals to be exchanged. Prokaryotes use conjugation as a means for horizontal gene transfer from one cell to another. Eukaryotes use gap junctions to permit passage of ions and small molecules between the cytoplasm of one cell to another. Many quorum sensing eukaryotes and prokaryotes use extracellular signaling molecules to synchronize their gene expression, behavior, and population density and thus exhibit multicellular properties [1],[2]. Other single-celled or multicellular organisms (and even cancerous cells) release extracellular membrane vesicles or exosomes to shuttle microRNAs, mRNAs, DNA fragments, and proteins to a recipient cell [3]. Additionally, recent research has shown nanotubes (also called tunneling nanotubes (TNTs) or membrane nanotubes (MNTs)) to transiently form between two or more eukaryotic or prokaryotic cells (Figure 1). These nanotubular channels are cell-to-cell plasma membrane connections of varying lengths that allow for bridging cargo exchange and, in some cases, multicellular properties.
Emerging research within the past 10 years has affirmed cellular nanotubes to be involved in complex signaling processes such as viral and prion pathogenesis among eukaryotic cells and metabolic or genetic exchanges among many types of bacteria. Surprisingly, the retrovirus HIV-1 has been shown to move between T-cells, macrophages, and B-cells by use of a nanotube-delivery mechanism for viral proteins [15],[16],[20],[22]. The leukemia-causing HTLV-1 similarly spreads throughout human T-cells by a similar mechanism to HIV-1, and this mechanism allows for a powerful and rapid transmission of the virus throughout the immune system. Prions (PrPSc) that cause degenerative neural disease have been shown to take over nanotubes in bone-marrow derived dendritic cells and spread by them to neural cells, allowing for a safe intercellular route to the brain [12]. Pathogens can indeed utilize already-existing nanotubes between cells, but they have been shown in some cases to even induce nanotube formation [22]. Elucidation of these eukaryotic mechanisms could provide potential targets for drug therapy in otherwise untreatable or hard-to-treat diseases. However, transport of cytoplasmic material through nanotubes is not an activity limited only to eukaryotic cells. Both similar and diverse species of bacteria have been observed to exchange not only cytoplasmic constituents, but also genetic material in the form of plasmids [19]. Dynamic nanotubes have been witnessed between the likes of B. subtilis, E. coli, Staphylococcus aureus, and Acinetobacter baylyi [18],[19]. The new discovery of a stress-induced mutualistic nutrient exchange between distantly related species of bacteria pushes the boundaries of our definition of “unicellular” organisms and prompts us to consider bacteria as a multicellular, interconnected domain of life [18].
Structure
Cellular nanotubes are membranous, tubular protrusions ranging anywhere from 50 nm to several hundred nm in diameter and, in some cases, extending hundreds of μm (several cell diameters) in length [4],[5]. First discovered by Rustom et al. 2004, these cylindrical channels were proposed to facilitate the traffic of intracellular vesicles and mediate membrane continuity between rat pheochromocytoma (PC12) cells [5]. PC12 cells were established from rat adrenals almost 30 years earlier as tools for neurobiological and neurochemical investigations [30]. In 2000, Rustom found strange structures connecting these rat tumor cells using fluorescence microscopy but thought of them only as plate scratches; four years later, he and his team were able to more accurately identify and characterize their morphology [10],[5]. In the same year, Önfelt et al. 2004 discovered nanotubes to transiently connect networks of immune cells [6]. As such, the mechanism for nanotube formation in eukaryotic animal cells has been largely characterized.
In human monocyte-derived macrophages, the cytoskeleton protein F-actin has been shown to support nanotubes of varying structure and function. These nanotubes also were shown to accommodate extracellular pathogens, such as the bacteria Mycobacterium bovis bacillus calmette-Guerin (BCG) that “surfed” along the surface of the nanotube and were subsequently phagocytosed by the macrophage [7]. Bridging between white blood cells, thin (<0.7 μm in diameter) nanotubes are composed of F-actin and allow for unidirectional movement of intracellular cargo within the nanotube [7]. Contrarily, wider (>0.7 μm in diameter) nanotubes composed of both actin and microtubules were found to aid in the bidirectional movement of organelles (lysosomes and mitochondria) and lipid vesicles between the macrophages [7]. This study by Önfelt et al. 2004 suggested a defensive functionality of nanotubes, which could bind extracellular bacteria and shuttle them toward the macrophage for phagocytosis.
Although F-actin was generally accepted to be a necessary component of eukaryotic nanotubes, Veranič et al. 2008 characterized novel type II nanotubes between RT4 and T24 urothelial cells that were long and stable due to a presence of cytokeratin and absence of F-actin. These type II nanotubes, which could extend up to several 100 μm in length, seemed to lack any signaling functionality but could connect cells that were in the process of dissociating for more than 2 hours [9]. Likewise, Wang et al. 2012 found nanotubes between neurons astrocytes that instead all contained microtubules—these nanotubes were actually longer if they lacked F-actin than if they were supported by it [8].
Formation
<http://link.springer.com/article/10.1007%2Fs11427-013-4548-3>.
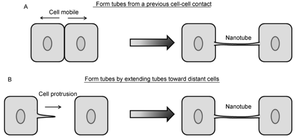
Contrarily, nanotubes are inhibited by chemical fixation, mechanical stress, and prolonged light exposure, making these structures intricately difficult to study [28]. However, through different methods, nanotubes have been found to form in two distinct general ways that differ per animal cell type (Figure 2) [10],[11]. One possibility is that actin polymerizes within a cell, causing a protrusion that can contact another cell. Such an approach would be useful for immobile or less-mobile cells like rat pheochromocytoma PC12 [10],[23]. Alternatively, cells that are in prolonged contact for a several minutes could move apart, trailing a newly formed nanotube between them in the separated space. This strategy is used by mobile cell types, such as T-cells that have an increased chance for collision with other cells [10],[15].
A diverse range of conditions has been found to contribute to nanotube formation in animal cells [25]. Blood serum starvation and lipopolysaccharide-induced inflammation has been shown to stimulate formation of nanotubes up to 330 μm long in mice cornea [26]. Oxidative cellular stress in the form of hydrogen peroxide has been shown to induce nanotubes between neural cells [21]. The interaction of the Fas ligand with its receptor and subsequent activation of Rho GTPase has also been shown to cause nanotube formation in T-cells; this mechanism is surprising, as the stimulation of the Fas ligand has been previously characterized to induce apoptosis [25],[27]. Interestingly, viral infection has been shown to induce nanotube formation. Although HIV-1 does not cause increased amounts of nanotubes on or between T-cells, it correlated with an increased number of nanotubes on human macrophages. Following a similar trend but with a different cell type, the retrovirus HTLV was found to stimulate a larger number of nanotubes on unactivated, static T-cells [15],[22].
Viral Pathogenesis
HIV
For viruses, the mammalian immune system presents a major obstacle. The best viral defense is to reside inside a host cell and replicate from within that cell—or, if possible, to spread intercellularly without ever abandoning cellular confines. Sowinski et al. 2008 and Eugenin et al. 2008 proposed such a mechanism for AIDS pathogenesis within the human immune system. In this model, the immunodeficiency virus HIV-1 infects either T cells or macrophages and exploits nanotubes to transmit the virus to other uninfected cells [15],[16]. In doing so, HIV could effectively evade extracellular immune response and increase its chance of reproducing infection [16]. In total, findings by these researchers provide a nascent framework for how HIV can effectively evade and destroy the human immune system by hiding within it.
Sowinski et al. 2008 looked for HIV-1 transmission via nanotubes connecting T-cells in the human immune system, witnessing nanotube formation only on T cells that adhered to a fibronectin coverslip. The team used recombinant HIV in which the open reading frame of the capsid-encoding Gag gene was fused with GFP, so that a Gag-GFP chimeric protein would be expressed within those T-cells. The fluorescent Gag was found to quickly move through nanotubes from one T-cell to another, validating the researchers’ hypothesis that nanotubes between T-cells could effectively function as an avenue for intercellular transmission of HIV [15].
In 2014, Lachambre et al. characterized nanotube structure of HIV infected T-cells to determine if cell membrane molecules involved in HIV-1 budding processes were also present in nanotubes [20]. They investigated the presence of the proteins HIV-1 Gag, which is involved in budding, and phosphatidylinositol(4,5) bisphosphate (PI(4,5)P2), which facilitates transport of Gag to the plasma membrane. In addition, they looked for tetraspanins, which colocalize and interact with Gag at the plasma membrane. Whereas the tetraspanin CD81 preferred no specific localization and was observed throughout the nanotube membrane, both Gag and PI(4,5)P2 were found to localize together in the nanotube tip that would initiate cell-to-cell contact. Furthermore, Lachambre et al. discovered that actin polymerization occurs at these tips by the presence of proteins that mediate membrane-actin interactions. These data suggest that we should view the T-cell nanotube tip as a separate domain in which HIV-1 localizes and from which HIV-1 buds off [20].
<http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2701345/>.
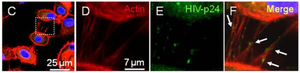
T-cell nanotubes were not the only means by which HIV could disseminate safely and intercellularly throughout the human immune system. Eugenin et al. 2008 searched for the HIV pathogenesis via nanotubes connecting human macrophages. When infected by HIV, macrophages are largely responsible for inducing brain tissue inflammation and dementia [16]. Strikingly, HIV infection correlated with a larger number of both small and large nanotubes formed between macrophages, implying that viral pathogenesis induces additional nanotube formation between macrophages. Within these nanotubes, the researchers detected the HIV p24 viral protein through immune staining (Figure 3) [16]. Eugenin et al. 2008 suggest that nanotubes may work in coordination with receptor-mediated HIV pathogenesis by CCR5 and CD4, in order to increase the chance for replicative success and viral communication between macrophages [16]. Moreover, Xu et al. 2009 discovered HIV transmission from macrophages to B-cells by detection of the HIV-1 protein Nef [22].
HTLV
The T-cell leukemia virus type 1 (HTLV-1) is a causative agent of human T-cell leukemia and lymphoma, and this retrovirus also causes tropical spastic paraparesis and HTLV-1-associated myelopathy [29]. This virus usually spreads via the virological synapse, but recent studies have implicated that not only does HTLV-1 transmit between T-cells through nanotubes, but it also increases the number of nanotubes per T-cell it infects, effectively increasing the opportunity for immobile, unactivated T-cells to exchange HTLV-1 [29].
In its viral genome, HTLV-1 expresses a protein called p8, which down-regulates T-cell receptor signaling. In 2010, Van Prooyen and colleagues discovered a previously uncharacterized mechanism for HTLV p8 transport and infection throughout nanotubes (called “conduits” in this literature) connecting T-cells. These nanotubes contained F-actin and connected T-cells from cytoplasm to cytoplasm, so they shared the same cytoskeletal and functional properties as other characterized nanotubes. Similarly to Sowinski et al. 2008, Van Prooyen et al. 2010 found that resting T-cells formed more nanotubes than activated ones [15],[29]. After observing that T-cells without the virus went on to obtain it after being cocultured with transfected with HTLV p8 T-cells, the researchers sought not only to find if nanotubes were responsible for this transmission, but also to discover if HTLV-1 prompted increased nanotube expression in infected cells.
As compared to non-infected control T-cells and T-cells infected with HTLV p12, HTLV p8 infected T-cells were found to increase the overall amount of cells containing nanotubes and the length of these nanotubes, suggesting that this p8 viral protein is sufficient by itself to induce nanotube formation in T-cells. Likewise, the percentage of T-cells with three or more nanotubes was increased upon HTLV p8 transfection. Unlike the discovery of nanotube-surfing bacteria by Önfelt et al. 2006, HTLV p8 was not found to travel on the surface of the nanotube; HTLV p8 actually spread upon contact from an infected T-cell to an uninfected T-cell by means of intracellular cytoplasmic extension. In doing so, HTLV-1 utilizes the same evolutionary tactics as HIV-1—residing and then rapidly disseminating within the immune system provides for a compelling viral survival strategy [29].
Prion Pathogenesis

Nanotubes have been found to aid in the transport of prions (PrPSc)—the causative agents of variant Creutzfeldt-Jakob disease—between eukaryotic neuronal and dendritic cells (Figure 4) [12]. In 2009, Gousset et al. proposed a mechanism for prion transport in nanotubes between bone-marrow derived dendritic cells and primary neurons, permitting intercellular transport of prion proteins toward the central nervous system [12]. The researchers found actin-containing nanotubes bridging between embryonic mice CAD cells, which are commonly used to study neuronal disease. Immunofluorescence revealed that these nanotubes allowed for free passage of prions between infected and non-infected cells. Moreover, the researchers used immunohistofluorescence to observe prion transport between a bone-marrow-derived dendritic cell and the axon on the CAD cell. Altogether, the findings by Gousset et al. 2009 suggest a rapid transport of prions throughout the body that is more specific and effective than that of exosomes [12]. The experiments also provide a potential pathway for prion pathogenesis, both within the brain and from the lymphatic system toward the brain. Also worth mention is the alternative hypothesis that, similarly to many retroviruses, prions may induce the formation of filopodia, which are actin-powered bridges that span from cell-to-cell. Unlike nanotubes, filopodia do not connect cytoplasm. However, they do allow for ligand exchange and viral transfer on the cell surface [13],[14].
Genetic and cytoplasmic exchange in B. subtilis
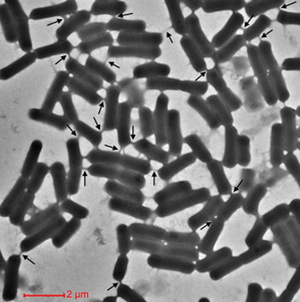
In 2011, Dubey et al. demonstrated that bacteria could share antibiotic resistance through nanotubes and even utilize nanotubes to anticipate environmental changes [19],[24]. The researchers found nanotubes to connect B. subtilus to Staphylococcus aureus and the evolutionarily distant E. coli and permit the molecular exchange of cytoplasmic protein and plasmid DNA between cells [19]. They visualized cytoplasmic molecules moving between adjacent B. subtilus cells on a solid surface with GFP and the dye calcein. To investigate the means by which these molecules were transferred, Dubey et al. 2011 used high-resolution scanning electron microscopy and found networks of nanotubes connecting the cells (Figure 1) [19]. The nanotubes were both large and small, ranging anywhere up to 1 μm in length and 30 to 130 nm in width. Interestingly, the team also found branched nanotubes that connected multiple B. subtilus cells together at once. All of these nanotubes were seemingly random in their placement on the cell surface, and they ceased to exist once the cells were suspended in liquid media. By thin section analysis, the researchers found the nanotubes to be composed of both cytoskeletal and cytoplasmic proteins [19].
Dubey et al. 2011 used two different antibiotic selection genes in coculture to demonstrate exchange of both non-hereditary and hereditary antibiotic resistance between B. subtilus. They used the proteins chloramphenicol acetyltransferase (Cat) and erythromycin resistance methylase (Erm), which confer resistance to chloramphenicol (Cm) and lincomycin (Lin), respectively. When two strains of B. subtilus, each containing chromosomally encoded resistance to either Cm or Lin, were mixed in co-culture on plates containing both Cm and Lin, the entire population of genetically varying cells survived. However, genetic analysis revealed no doubly resistant genotype in either cell that survived in this population, suggesting not a permanent incorporation of DNA but a bidirectional, transient resistance via antibiotic transfer. While the prospect of chromosomal DNA transfer seemed unlikely, Dubey et al. 2011 used another antibiotic selection experiment and found neighboring cells capable of exchanging extrachromosomal genetic information in the form of nonconjugative plasmids. To discount the possibility of non-hereditary resistance, second-replica plates were shown to also grow on antibiotics. This indicated that in addition to transferring cytoplasmic proteins, nanotubes could also grant movement of genetic material [19].
Remarkably, Dubey et al. 2011 observed nanotube formation between the two firmicutes B. subtilus and Staphylococcus aureus and even between the gram positive B. subtilus and the distant, gram negative proteobacteria E. coli. Additionally, nanotubes between S. aureus and E. coli were found, indicating a large scale bacterial commonality in this functional feature. They used high-resolution scanning electron microscopy to visualize nanotubes between these species, and they assumed that cytoplasmic elements would also have been transferred. However, they observed that nanotubes formed by E. coli cells were much thinner than those formed by B. subtilus or S. aureus, which may suggest that nanotubes differ among gram positive and negative bacteria. With these findings, Dubey et al. discuss the pathogenic implications of bacterial nanotubes and propose that pathogenic bacteria could use nanotubes to transfer virulence to non-pathogenic bacteria [19].
Metabolic mutualism between E. coli and Acinetobacter baylyi
Previous research in neural cells has found stress, in the form of hydrogen peroxide addition, to be a sufficient nanotube inducer, as a stressed cell always initiated nanotube connection with an unstressed cell [21]. However, the question of nanotube significance in nutrient exchange has, up until recently, remained unclear. Through a study of cross-feeding interactions, Pande et al. 2015 found environmental stress, in the form of essential amino acid knockout, to sufficiently induce nanotube formation between starving gammaproteobacterial cells that could complement each other’s metabolic needs [18]. This differed from Dubey et al. 2011, which found that nanotube count between B. subtilus cells actually lowered on minimal media [19]. In their experimental design, Pande et al. 2015 created auxotrophic samples by knocking out genes encoding for histidine and tryptophan biosynthesis (ΔhisD and ΔtrpB) in E. coli and Acinetobacter baylyi strains. In two additional samples, the researchers deleted genes encoding for proteins that regulate these biosynthesis pathways, conversely resulting in E. coli and A. baylyi strains that overproduced histidine and tryptophan (ΔhisL and ΔtrpR). The team discovered contact-dependent cross feeding between cocultures that were both lacking yet also overproducing complementary amino acids. For example, an E. coli sample lacking tryptophan but overproducing histidine compensated by connecting a nanotube with an A. bayli sample lacking histidine but overproducing tryptophan. While E. coli was able to initiate nutrient exchange with cells of its own species, A. baylyi was not. Moreover, they discovered through fluorescent tagging that this exchange of amino acids occurred bidirectionally. However, unlike Dubey et al., an ampicillin selection experiment showed no evidence of plasmid DNA transfer between E. coli and E. coli or A. baylyi [18].
Once Pande et al. 2015 supplied the liquid growth medium with histidine and tryptophan, nanotube formation ceased between all bacteria. Therefore, compared to samples separated by a filter that allowed for diffusion of nutrients but did not allow for cellular contacts, a nutrient-deprived environment was said to induce nanotube formation and encourage the sharing of nutrients and cytoplasmic constituents between stressed cells. Once the nutrient-depleted environment was established, however, Pande et al. 2015 were still uncertain if nanotube formation between the cells was specifically chemosensory or incidentally promiscuous. Nonetheless, the team reports their findings as crucial toward our current understanding of bacterial ecology. They found distant relatives of bacterial cells that were able to mutually benefit one another chemically while still maintaining their own genetic integrity, implying either a physiological interconnectedness or even multicellular characteristic between the bacterial domain as a whole [18].
References
1. Waters, C. M., and Bassler, B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346. doi: 10.1146/annurev.cellbio.21.012704.131001 <http://www.annualreviews.org/doi/pdf/10.1146/annurev.cellbio.21.012704.131001>.
2. Montgomery, K.; Charlesworth, J.C.; Lebard, R.; Visscher, P.T.; Burns, B.P. Quorum Sensing in Extreme Environments. Life 2013,3, 131–148. <http://www.mdpi.com/2075-1729/3/1/131/htm>.
3. Kahlert C, Kalluri R (2013) Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl) 91: 431–437. <http://link.springer.com/article/10.1007/s00109-013-1020-6/fulltext.html>.
4. K. Schara, V. Janša, V. Šuštar et al., Mechanisms for the formation of membranous nanostructures in cell-to-cell communication. Cellular and Molecular Biology Letters, vol. 14, no. 4, pp. 636–656, 2009. <http://www.degruyter.com/view/j/cmble.2009.14.issue-4/s11658-009-0018-0/s11658-009-0018-0.xml>.
5. Rustom A, Saffrich R, Markovic I, Walther P, Gerdes HH. Nanotubular highways for intercellular organelle transport. Science 2004; 303: 1007–1010. <http://www.sciencemag.org/content/303/5660/1007.long#F1>.
6. Önfelt B, Nedvetzki S, Yanagi K, Davis DM: Cutting edge: Membrane nanotubes connect immune cells. J Immunol 2004, 173:1511-1513. <http://www.jimmunol.org/content/173/3/1511.long>.
7. Önfelt B, et al. Structurally distinct membrane nanotubes between human macrophages support longdistance vesicular traffic or surfing of bacteria. J Immunol 2006;177:8476–8483. <http://www.jimmunol.org/content/177/12/8476.long>.
8. Wang X, Bukoreshtliev NV, Gerdes HH. Developing neurons form transient nanotubes facilitating electrical coupling and calcium signaling with distant astrocytes. PLoS One 2012; 7:e47429. <http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0047429>.
9. Veranic P, Lokar M, Schutz GJ, Weghuber J, Wieser S, et al. Different types of cell-to-cell connections mediated by nanotubular structures. Biophys J 2008, 95:4416–4425. <http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2567924/>.
10. Zhang JiangHui, Zhang YouYi. Membrane nanotubes: novel communication between distant cells. Life Sciences 2013, 10.1007/s11427-013-4548-3. <http://www.ncbi.nlm.nih.gov/pubmed/24008389 >.
11. Davis DM, Sowinski S Membrane nanotubes: dynamic long-distance connections between animal cells. Nat Rev Mol Cell Biol 2008, 9:431–436. <http://www.nature.com/nrm/journal/v9/n6/full/nrm2399.html>
12. Gousset K et al. Prions hijack tunnelling nanotubes for intercellular spread. Nat Cell Biol 2009 11:328–336. <http://www.nature.com/ncb/journal/v11/n3/full/ncb1841.html>.
13. Gousset K, Zurzolo C. Tunnelling nanotubes: a highway for prion spreading? Prion 2008, 3:94–98. <http://www.tandfonline.com/doi/abs/10.4161/pri.3.2.8917#.VTM9GJTF-5I>.
14. Sherer NM, Mothes W (2008) Cytonemes and tunneling nanotubules in cell-cell communication and viral pathogenesis. Trends Cell Biol 18:414–420. <http://www.ncbi.nlm.nih.gov/pubmed/18703335>.
15. Sowinski S, et al. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nature Cell Biol 2008;10:211–219. <http://www.nature.com/ncb/journal/v10/n2/full/ncb1682.html>.
16. Eugenin EA et al. Tunneling nanotubes (TNT) are induced by HIV-infection of macrophages: a potential mechanism for intercellular HIV trafficking. Cell Immunol 2009, 254:142–148. <http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2701345/>.
17. Eugenin EA et al. Tunneling nanotubes (TNT): a potential mechanism for intercellular HIV trafficking. Commun Integr Biol 2009. 2:243–244. <http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2717534/>.
18. Pande, S. et al. Metabolic cross-feeding via intercellular nanotubes among bacteria. Nat. Commun. 6, 2015. 6238. <http://www.nature.com/ncomms/2015/150223/ncomms7238/abs/ncomms7238.html>.
19. Dubey GP and Ben-Yehuda S. Intercellular nanotubes mediate bacterial communication. Cell. 2011. 144: 590–600. <http://www.sciencedirect.com/science/article/pii/S009286741100016X>.
20. Lachambre S, Chopard C, Beaumelle B. Preliminary characterisation of nanotubes connecting T-cells and their use by HIV-1. Biol Cell 2014; 106: 394-404. <http://onlinelibrary.wiley.com/doi/10.1111/boc.201400037/epdf>.
21. Wang, Y., Cui, J., Sun, X. and Zhang, Y. (2011) Tunneling-nanotube development in astrocytes depends on p53 activation. Cell Death Differ. 18, 732–742. <http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3131904/>
22. Xu W, Santini PA, Sullivan JS, He B, Shan M, Ball SC. HIV-1 evades virus-specific IgG2 and IgA responses by targeting systemic and intestinal B cells via long-range intercellular conduits. Nat Immunol 2009; 10:1008 - 1017. <http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2784687/>.
23. Bukoreshtliev NV, Wang X, Hodneland E et al. Selective block of tunneling nanotube (TNT) formation inhibits intercellular organelle transfer between PC12 cells. FEBS Lett 2009; 583: 1481–1488. <http://www.ncbi.nlm.nih.gov/pubmed/19345217>.
24. Ficht, T. Bacterial exchange via nanotubes: lessons learned from the history of molecular biology. Front, Microbiol 2008. 10.3389/fmicb.2011.00179. <http://journal.frontiersin.org/article/10.3389/fmicb.2011.00179/abstract>
25. Kimura S, Hase K, Ohno H. The molecular basis of induction and formation of tunneling nanotubes. Cell Tissue Res, 2012. 10.1007/s00441-012-1518-1. <http://link.springer.com/article/10.1007%2Fs00441-012-1518-1>
26. Chinnery, H.R., E. Pearlman, and P.G. McMenamin. 2008. Cutting edge: Membrane nanotubes in vivo: a feature of MHC class II+ cells in the mouse cornea. J. Immunol. 180:5779–5783. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3392179/
27. Luchetti et al. Fas Signaling Promotes Intercellular Communication in T Cells. PLoS One, 2012. e35766. doi:10.1371/journal.pone.0035766. <http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0035766>.
28. Wang, X., and Gerdes, H.-H. Long-distance electrical coupling via tunneling nanotubes. Biochim. Biophys. Acta. 2011. PMID:21930113. <http://www.sciencedirect.com/science/article/pii/S0005273611003087>.
29. Van Prooyen N, Gold H, Andresen V, Schwartz O, Jones K, Ruscetti F, et al. Human T-cell leukemia virus type 1 p8 protein increases cellular conduits and virus transmission. Proc Natl Acad Sci USA. 2010;107:20738–43. <http://www.pnas.org/content/107/48/20738.long>.
30. Greene, LA and Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci USA. 1976 Jul; 73(7): 2424–2428. <http://www.ncbi.nlm.nih.gov/pmc/articles/PMC430592/>.
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2015, Kenyon College.
