Neorickettsia sennetsu
A Microbial Biorealm page on the genus Neorickettsia sennetsu
Classification
Higher order taxa
Kingdom: Bacteria
Phylum: Proteobacteria
Class: Alphaproteobacteria
Order: Rickettsiales
Family: Anaplasmataceae
Genus
Genus: Neorickettsia
Species: sennetsu
|
NCBI: Taxonomy |
Description and significance
When research began on N. sennetsu it was classified as Ehrlichia sennetsu, based on the morphological, ecological, epidemiological and clinical infection characteristics, which were similar to other Erhlichia spp. More recent genetic analysis of the 16S rRna genes, heat shock proteins (groESL), surface proteins, and most recently RNA polymerase beta-subunit have reclassified Ehrlichia sennetsu as Neorickettsia sennetsu. The Anaplasmataceae family now consists of four genera, Ehrlichia, Neorickettsia, Wolbachia and Anaplasma.
Members of the family Anaplasmataceae are Gram-negative obligatory intracellular bacteria, meaning that they cannot survive out side their eukaryotic host cell. The different species within this family are specific to different compartments with in the host cell, such as granulocytes, platelets, endothelial cells, monocytes, macrophages, or red blood cells (9).
N. sennetsu is only infectious to human beings. This disease has only been found in Japan and Malaysia3. Though unknown, the vector of N. sennetsu is thought to be trematodes in fish. The bacteria infect those that eat improperly cooked fish or raw fish (according to the CDC). The bacteria invade monocytes and macrophages. The infection causes Sennetsu fever, which has the following symptoms fever, chills, headache, sore throat, insomnia (4).
The entire genome of all four known strains of N. sennetsu are sequenced.
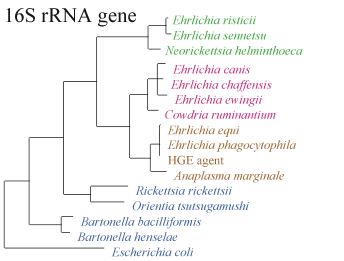
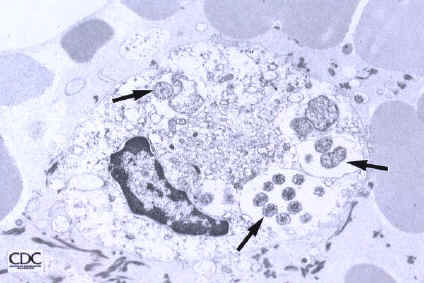
The following shows (a)the relationship between different species by the sequence of the 16S RNA gene and (b) the infection of a cell by E. sennetsu(5):
Note: The above pictures are displayed on behalf of the Center for Disease Control
Genome structure
Total number of DNA molecules: 1; circular chromosome; no plasmids
Total size of all DNA molecules: 859006bp
Total genes: 971
Protein coding genes: 935
tRNA genes: 33
mRNA genes: 3
Cell structure and metabolism
Presence of flagella: No
Interaction: Animal pathogen in Humans
Number of membranes: 2
Number of inteins: 0
Vector: believed to be Trematodes
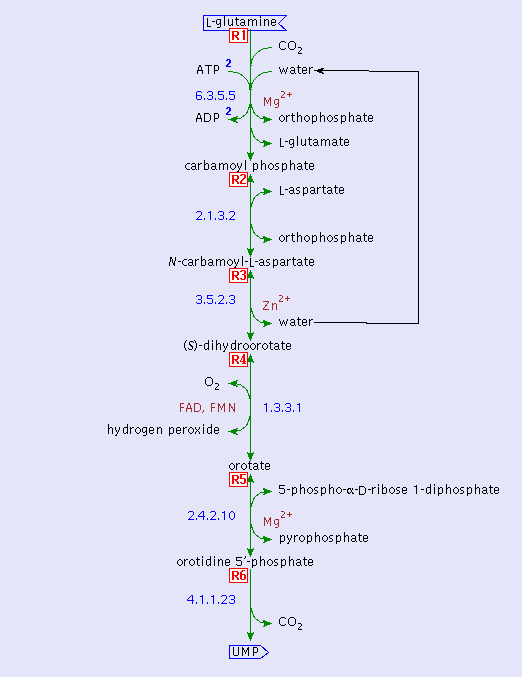
Glutamine (not glutamate) is the preferred substrate for ATP synthesis for in vitro cultures of N. sennetsu. Difficulties analyzing metabolic preoperties arise from the bacteria not being able to survive out of the host cell. Biological techniques are not advanced enough to fully study how N. sennetsu acquires energy and nutrients.
The following is the metabolism of glutamine by N. sennetsu:
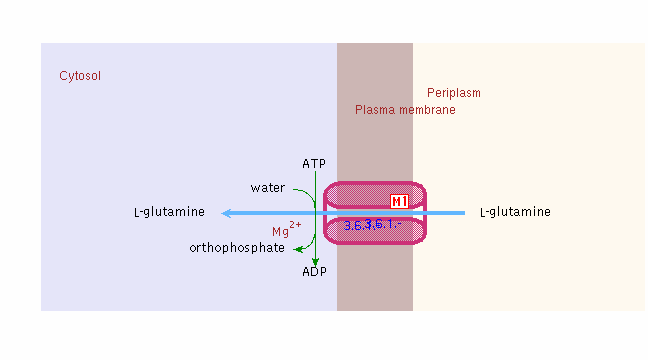
Below is the transport of glutamine into the bacterial infected vacuole of the host cell:
Pathology
The vector for N. sennetsu is thought to be trematodes in fish. Ingestion of the vector through eating raw fish or improperly cooked fish can lead to infection. “Infection with E. sennetsu results in an acute febrile illness with lymphocytosis and postauricular and posterior cervical lymphadenopathy similar to mononucleosis3.” Sennetsu fever causes enlargement of the lymph nodes, liver and spleen. The symptoms are non-specific flu-like symptoms, such as fever, chills, headache, sore throat, and insomnia.
The infection is usually mild to moderate and can be treated with tetracyclines, specifically doxycycline for 10 days. Along with tetracyclines, the bacteria is susceptible to ciprofloxacin and rifampin. N. sennetsu has been tested against more common drugs, but was found to be resistant to erythromycin, chloramphenicol, penicillin (usually used against Gram-positive bacterial infections), gentamicin, and co-trimoxazole.
N. risticii, N. sennetsu, SF agent, and N. helminthoeca are antigenically cross-reactive. The current test for N. sennetsu infection is through antibody titer against Ehrlichia canis, which is canine ehrlichiosis (3). N. risticii causes Potomac horse fever. Research has been done on horses that are first injected with N. sennetsu. These horses do not develop Sennetsu, as the bacteria only cause disease in humans. The horses had high levels of immunoglobulin G antibody, which reacted against injections of N. risticii, protecting the horse from disease. Through immunofluorescent antibody testing, the immunoglobulin G antibody reacted against both E. sennetsu and E. risticii antigens (11).
When the bacteria interacts with a host cell, many signals are created in the host cell and the bacteria. The bacteria attach to different receptors on the outside of the cell, L-selectin and E-selectin, which causes the inhibition of phagolysosomal fusion, apoptosis, and JAK/STAT activation, and downregulates IL-12, IL-15, IL-18, TLR2 and 3, and CD14 (10). The signals “disarm” the host cell alarm system against infection by pathogens, allowing the entry of the bacteria to the host cell. These signals also condition the cell for sharing space and nutrients with the new bacteria and disguising them as “insiders”(9).
Current Research
1. Extensive research on the heat shock proteins HSP60 and HSP70 and their role in E. sennetsu is currently under way. The promoters for the heat shock proteins (HSP) become active in situations when the bacteria live in a higher temperature environment than usual, such as when E. sennetsu infects a human. The similarities between E. sennetsu heat shock proteins and those of other better-known bacteria lead to the belief that the E. sennetsu HSP60 and HSP70 proteins have the same functions. The promoter region for the heat shock 32 sequences lie upstream of the typical 35 and 10 promoter consensus of the 70. This heat shock promoter region is analogous with the 35 promoter region of E. coli heat shock 32 RNA polymerase. In several different and more studied bacteria, including Mycobacterium, Borrelia, Chlamydia, and Legionella spp., the HSP have been recognized by the immune system as common antigens in its response to bacterial infection. Recently, studies have shown bacterial HSP60 and HSP70 to “modulate immunity by directly inducing cytokine mRNA production in macrophages”. HSP70 have also been shown to mediate the role of pathogen and host cell binding. This interaction between host cell and pathogen is critical for N. sennetsu infection because it cannot survive outside host cells. Further research needs to be done to understand the specific role heat shock proteins have in increasing the viability of the bacteria when under thermal stress (2).
2.New easier ways to test for the infection of N.sennetsu, and other erhlichiosis diseases in under way. Current diagnosis using serology or PCR is not sufficient in establishing the “true impact of this emerging virus-like illness” (10). Citrate synthase gene sequences are thought to be a way to diagnosis a patient with ehrlichiosis of some type and then be able to correctly identify the species.
3.Genera of the Anaplasmataceae family are under scrutiniy to define them by the correct taxonomy. Much research is going into different methods for defining and comparing the involved species. New methods include GroEL (heast shock protein) amino acid sequences, RNA polymerase beta-subunit sequences and citrate synthase gene sequence (12, 13).
4.Analysis of the immunodominant outer membrane proteins are being studied to understand the bacteria’s antigenic variability (14). Few antigenic markers have been found for this species, one of which is P51. The four strains of N. sennetsu have identical p51 sequences, leading scientist to believe this gene is important to the survival of the bacteria (15).
References
(1)http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?org=ges
(2)Zhang, Y., Ohashi N., and Rikihisa Y. “Cloning of the Heat Shock Protein 70 (HSP70) Gene of Ehrlichia sennetsu and Differential Expression of HSP70 and HSP60 mRNA after Temperature Upshift”. Infect Immun. July 1998. Vol. 66, No. 7. p. 3106-3112.
(3)Tachibana N. “Sennetsu fever: the disease, diagnosis, and treatment”. In: Leive L, ed. Microbiology--1986. Washington, DC: American Society for Microbiology, 1986:205-8
(4)Travelers’ Health: Yellow Book. Health Information for International travel, 2005-2006, CDC.
(5)Walker, D., Dumler, J. “Emergence of the Ehrlichioses as Human Health Problems”. CDC. http://www.cdc.gov/ncidod/dvrd/ehrlichia/Organisms/Organism.htm
(6) Sazanov, L.A., Hinchliffe, P. “Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus”. Science. 2006. p. 311:1430-6.
(7) Hinchliffe, P., Carroll, J., Sazanov, L.A. “Identification of a novel subunit of respiratory complex I from Thermus thermophilus”. Biochemistry. 2006. p. 45:4413-20.
(8)PUMA2--grid-based high-throughput analysis of genomes and metabolic pathways. Maltsev N, Glass E, Sulakhe D, Rodriguez A, Syed MH, Bompada T, Zhang Y, D'Souza M. EMP Metabolic reconstruction for: Neorickettsia sennetsu str. Miyayama 222891.
(9)Rikihisa Y. “Mechanisms to create a safe haven by members of the family Anaplasmataceae”. Ann N Y Acad Sci. 2003 Jun;990:548-55.
(10) Walker, D.H. “Ehrlichia under our noses and no one notices”. Arch Virol Suppl. 2005;(19):147-56.
(11) : Rikihisa Y, Pretzman CI, Johnson GC, Reed SM, Yamamoto S, Andrews F.Infect Immun. “Clinical, histopathological, and immunological responses of ponies to Ehrlichia sennetsu and subsequent Ehrlichia risticii challenge.” 1988 Nov.,56(11):2960-6.
(12) Yu XJ, Zhang XF, McBride JW, Zhang Y, Walker DH. “Phylogenetic relationships of Anaplasma marginale and 'Ehrlichia platys' to other Ehrlichia species determined by GroEL amino acid sequences.” Int J Syst Evol Microbiol. 2001 May;51(Pt 3):1143-6.
(13) Taillardat-Bisch AV, Raoult D, Drancourt M. “RNA polymerase beta-subunit-based phylogeny of Ehrlichia spp., Anaplasma spp., Neorickettsia spp. and Wolbachia pipientis.” Int J Syst Evol Microbiol. 2003 Mar;53(Pt 2):455-8
(14) Hotopp JC, Lin M, Madupu R, Crabtree J, Angiuoli SV, Eisen J, Seshadri R, Ren Q, Wu M, Utterback TR, Smith S, Lewis M, Khouri H, Zhang C, Niu H, Lin Q, Ohashi N, Zhi N, Nelson W, Brinkac LM, Dodson RJ, Rosovitz MJ, Sundaram J, Daugherty SC, Davidsen T, Durkin AS, Gwinn M, Haft DH, Selengut JD, Sullivan SA, Zafar N, Zhou L, Benahmed F, Forberger H, Halpin R, Mulligan S, Robinson J, White O, Rikihisa Y, Tettelin H. “Comparative genomics of emerging human ehrlichiosis agents”. Epub 2006 Feb 17.
(15) Rikihisa Y, Zhang C, Kanter M, Cheng Z, Ohashi N, Fukuda T. “Analysis of p51, groESL, and the major antigen P51 in various species of Neorickettsia, an obligatory intracellular bacterium that infects trematodes and mammals.” J Clin Microbiol. 2004 Aug;42(8):3823-6.
Edited by student of Rachel Larsen and Kit Pogliano