Nitrosococcus oceani
A Microbial Biorealm page on the genus Nitrosococcus oceani
Classification
Higher taxa: Bacteria (domain); Proteobacteria (phylum); Gammaproteobacteria (class); Chromatiales (order); Chromatiaceae; (1) Species: Nitrosococcus oceani
Description and Significances
Nitrosococcus oceani is is a marine, gram-negative, chemolithoautotrophic, ammonia-oxidizing bacterium (AOB). Its main source of energy is from oxidizing ammonia. In return the cells release nitrogen and nitrogen oxide. Each cell can produce up to 2x10-6microM of NO2- /day (Watson 1962). It was the first nitrifying bacteria isolated from open ocean water by Watson in 1962. At the time it was known as Nitrosocystis oceanus. N.oceani is able to form cysts and zoogloea in harsh conditions. However, when its genome was completely sequence, little of its genome contributes to tolerate difficult environments (Klotz 2006). It was found in marine world. It was found in Pacific Ocean, and Atlantic Ocean. Oddly, it was found it also to be in the saline waters of Lake Bonney while using FISH for detection. It is widely distributed (Ward 2002). However, within the marine ammonia-oxidizing bacteria, it is dominant by betaproteobacteria (Nold 2000). Nold has used primers, which are complementary to the conservative region of amo and pmo gene, to detect the existence of both gammaproteobacteria and betaproteobacteria in marine water samples collected through out the world. It genome has been completely sequence by Klotz in 2006. In the ocean, it is a frequent target for bacterriophage.
While betaproteobacterial nitrifer has one rrn operon, N.oceani contains two complete sets of rrn operons. However, only one copy of the operon is required. Its genome encodes twenty aminoacyl-tRNA synthetases. It also encodes for two forms of LysRS and two distinct forms of GlxRS but AsnRS and GlnRS are not found. Found to have several sodium/hydrogen antiporters which plays an important role in maintaining intracellular pH allowing it to survive in high salt environment (Klotz 2006).
Its significances are that N. oceani has both nitrification and denitrification activities. It contributes significantly in denitrifying the toxic ammonium, a waste product of animals and plants, into nitrate releasing back into the atmosphere as a resource for plants to grow on. It also could denitrify NO3- back into N2 green gas. (4)
Background
In 1971, Watson classified ammonia-oxidizing bacteria into four genera based upon morphology, fine structure, and percentage of guanine-cytosine (GC) within genome of 27 strains of nitrying bacteria. Nitrosomonas and Nitrosocystis species form one group, both of them belong to gammaproteobacteria. Nitrosolobus and Nitrosospira form another group; both of these belong to betaproteobacteria. Later this classification wass confirmed by 16S rRNA sequence (Head 1993, Purkhold 2000). The sequence of amo and pmo genes are also utilized to build the phylogenetic tree (Purkhold 2000)
Nitrosococcus halophilus is the closet related species to Nitrosococcus oceani. Among other cultivated aerobic AOB, N.oceani and N. halophilus are the only two organisms that are gammaproteobacterial AOB, the rest are Betaproteobacteial AOB. Nitrosococcus oceani resides in saline environment, found in marine world or seawater. Its presence is detected using immunofluoresence and FISH. (Klotz 2006))
Structure and Metabolism
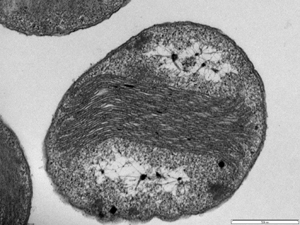
N.oceani's morphology ranges from spherical to ellipsoidal. Cells are 1.8-2.2 micrometer in diameter. Its cell wall is as thick as 250A while the plasma membrane's thickness is 80A. It is found as diploids, or occasionally as tetrads, and has flagella and two layers of membrane (Watson 1962). Within the cell there is a dense enveloping layer making up inner triplet structure; there is a series of flattened membrane-bound organelle lined up in the middle of the cell .
Nitrosococcus oceani relies on oxidizing ammonia to nitrite to obtain energy for growth. However, being a member of Chromatiaceae, N.oceani also participate in the sulfur cycle; it reduces sulfate into H2S (Klotz 2006).
As ammonia is transported into the cell, it passed by the membrane bound hertero-trimeric copper ammonia monooxygenase (AMO) enzyme to be oxidized to hydroxylamine (Hooper, 2005). The uptake of ammonia is regulated by the ratio of glutamine and glutamate (Klotz, 2006). Hydroxylamine is then oxidized to nitrite by periplasmic hydroxylamine oxidoreductase (HAO), which binds to seven c-type hemes and an active site heme, heme P460 (Numata 1990). HAO is also called octaheme HAO. The hydroxylamine which is missed by HAO would be oxidized by periplasmic monoheme cytochrome P460 protein (Vannelli 1996). The process of oxidizing is also found in the closely related genera Nitrosomonas and Nitrosospira of Betaproterobacteria (Head 1993). It is studied extensively in Nitrosomonas europaea. The process of oxidizing ammonia is done by following the flow of the electron. The electron obtained by octaheme HAO is transferred to membrane-bound ubiquinone mediated by tetraheme cytochrome c554 and membrane-associated tetraheme cytochrome cm552. Finally two electrons are fed into AMO and two into electron transport chain to cytochrome oxidase. The drawn-out steps of the process of oxidizing ammonia is supported by the clustering of hao genes in N. europaea (Bergmann, 1994). And Bergmann argues later in 2005 that the clustering of hao genes, with the backup of phylogenetic analyses, evolved in the gammaproterobacterial ancestors of extant autotroph ammonia oxidizing bacteria. The hao gene cluster found residing in betaproteobacterial aAOB as a result of horizontal gene transfer.
Genome
A complete sequence of amo operon, the amoCAB, for N. oceani has been completed by Alzerreca JJ. From the report the following is concluded: 49-53% is identical to the pmo gene encoding for mathane monooxygenase of methylococcus capsulatus, 39-42% identical to the amo gene of beta-proteobacteria. Within amo operon, three genes were found, the amoC, amoA, and amoB. There are 286bp and 65bp intergenetic spacer between amoC and amoA, between amoA and amoB relatively. Alzerreca concluded from the complete sequence of amo operon that this allow a finer phylogenetic sorting than 16S rRNA alone (Alzerreca, 1999)
Nitrosococcus oceani genome wass completed sequence in early 2006. N. oceani has one single circular chromosome 3,481,691 bp in size and one plasmid 40,420 bp in size. The chromosome contains 3,052 protein-encoding sequences while the plasmid encodes for 41 proteins (Klotz 2006). At least twenty-two genes codea for proteins which are involved in iron transport. In its genome can also found several uptake systems for other inorganic ions, several sodium/hydrogen antiporters, several protein export, secretion systems including a preprotein translocase, TatABC system, type II general secretion/pilus synthesis pathway, as well as type IV conjugal DNA/protein transfer system. It also encodes for form I RuBisCO It has a cluster of genes encoding for parts of PTS-type sugar transport system. Two large clusters of genes encode for flagellation and motility genes. Although it is able to metabolize methane, it is very limited because it has only one methyl-accepting chemotaxis protein (Klotz 2006).
Transportation
Nitrosococcus oceani genome size is 3.4Mb. Each Mb genome encodes for roughly 35.23 proteins for transportation making up a total of 124 proteins for the whole transportation system. N.oceani' primary transport relies on secondary transportation, secondary is ATP-dependent transportation. 50% of the total transporter proteins are secondary transporter, and 34.7% of the total transporter proteins contributes to ATP-dependent channels. Left with 7.3%, 4%, 4% encode for ion channels, phosphotransferase system, relatively. 4% of the total transporter proteins remained unclassifed.
Ecology
Nitrosococcus oceani impact on ecosystem is the massive release of nitrogen oxide and nitrogen gases into the atmosphere completing the nitrification cycle. Also it contributes to maintain the concentration of nitrate in the nitrogen pool in the deep ocean at 40 µM (Klotz 2006).
In the brackish Ganzirri lake, a correlation of the increasing of particulate organic carbon (POC), particulate organic nitrogen (PON) and the abundance of Nitrosococcus species is found positively (Zaccone 1996). However, a negative correlation was found with oxygen concentration. Interestingly, via immunofluorescent it was reported that there is a overlapping of polyclonal antiserum with rabbits antiserum. These polyclonal antisera were found to be cross-reacted with other ammonia-oxidizing strains, but not with other marine bacteria (Zaccone 1996).
Pathology
There is no known pathology caused by Nitrosococcus oceani or by related strains.
Application to biochemically
Not currently found to be useful for the development of biochemical technology.
Current Research
1.Klotz's laboratory at University of Louisville, which has recently completed sequencing the genome of Nitrosococcus oceani, now aims to study the metabolism pathway of ammonia catabolism and urea hydrolysis. Its project is to identify all the enzymes and conditions involving in the nitrification process.
2.Bess B. Ward's laboratory at Princeton University is doing a new protect on “Control of Denitrification in a Permanently Ice Covered antarctic Lak: Potential for Rgulation by Bioactive Metals”. The project main study site Taylor Valley of East Antarctica and Lake Bonney, where N. oceani were found. They traced metal distribution in the ecosystem which they think is related to the microbial activities. They focused on the impact of the availability of metal on microbial organisms.
References
1.Alzerreca JJ, Norton JM, Klotz MG. 1999. The amo Operon in Marine, Ammonia-oxidizing Gamma-proteobacteria.
2.Bergmann, D. J., D. A. Arciero, and A. B. Hooper. 1994. Organization of the hao gene cluster of Nitrosomonas europaea: genes for two tetraheme c cytochromes. J. Bacteriol.
3.Bergmann, David J., Hooper, Alan B., and Klotz, Martin G. 2005. Structure and Sequence Conservation of hao Cluster Genes of Autotrophic Ammonia-Oxidizing Bacteria: Evidence for Their Evolutionary History
4.Head, I. M., W. Hiorns, T. Embley, A. McCarthy, and J. Saunders. 1993. The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal RNA gene sequences. J. Gen. Microbiol. 13:1147-1153
5.Hooper, A. B., D. M. Arciero, D. Bergmann, and M. P. Hendrich. 2005. The oxidation of ammonia as an energy source in bacteria in respiration, vol. 2. Springer, Dordrecht, The Netherlands.
6. KlotzMartin G., Arp, Daniel J., Chain,Patrick S. G., El-Sheikh, Amal F., Hauser,Loren J., Hommes,G., Larimer, Frank W.,. Malfatti, Stephanie A, Norton,Jeanette M., Poret-Peterson, Amisha T., Vergez , Lisa M., and Ward, Bess B. 2006. Complete Genome Sequence of the Marine, Chemolithoautotrophic, Ammonia-Oxidizing Bacterium Nitrosococcus oceani ATCC 19707
7. Nold, Stephen C.,Zhou, Jizhong, Devol, Allan H., and Tiedje James M. 2000. Pacific Northwest Marine Sediments Contain Ammonia-Oxidizing Bacteria in the Beta Subdivision of the Proteobacteria
8.Numata, M., T. Saito, T. Yamazaki, Y. Dukumori, and T. Yamanaka. 1990. Cytochrome P-460 of Nitrosomonas europaea: further purification and further characterization. J. Biochem.
9.Paulsen, Ian T. Transporter Proteins in Nitrosocuccus oceani at www.membranetransport.org
10.Purkhold, U., A. Pommerening-Roser, S. Juretschko, M.C. Schmid, H.-P. Koops, and M. Wagner. 2000. Phylogeny of All Recognized Species of Ammonia Oxidizers Based on Comparative 16S rRNA and amoA Sequence Analysis: Implications for Molecular Diversity Surveys.
11.Vannelli, T., D. Bergmann, D. M. Arciero, and A. B. Hooper. 1996. Mechanism of N-oxidation and electron transfer in the ammonia oxidizing autotrophs, p. 80-87. In M. E. Lidstrom and F. R. Tabita (ed.), Microbial growth on C1 compounds. Kluwer Academic Publishers, Amsterdam, The Netherlands.
12.Watson, Stanley W. 1962. Characteristics of a Marine Nitrifying Bacterium, Nitrosocystis Oceanus Sp.N.
13.Watson, Stanley W., and Mandel Manley. 1971. Comparison of the Morphology and Deoxyribonucleic Acid Composition of 27 Strains of Nitrifying Bacteria.
14.Ward, Bess B., O'Mullan, Greogory D. 2002. Worldwide Distribution of Nitrosococcus oceani, a Marine Ammonia-Oxidizing Gammaproteobacterium, Detected by PCR and Sequecing of 16S rRNA and amoA Genes
15.Zaccone, R., G. Caruso, and M. Azzaro. 1996. Detection of Nitrosococcus oceanus in a Mediterranean Lagoon by Immunofluorescence
Edited by Khai Quach, student of Rachel Larsen at UCSD
KMG