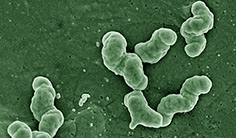
Nitrospira moscoviensis
Classification
Domain: Bacteria; Phylum: Nitrospirae; Class: Nitrospira; Order: Nitrospirales; Family: Nitrospiraceae
Species
NCBI: Nitrospira moscoviensis [1]
Nitrospira moscoviensis
Description
Nitrospira moscoviensis is a Gram-negative, non-motile, facultative aerobe. It is helical- and vibroid-shaped, but it often forms spirals. Cells are 0.9–2.2 µm × 0.2–0.4 µm in size and tend to form biofilms. They grow optimally at 39°C and require a pH between 7.6-8.0 [1]. N. moscoviensis is a member of the phylum Nitrospira, which is the largest known class of nitrogen-oxidizing bacteria [2]. It is the primary nitrite-oxidizer in soils, water treatment plants, and activated-sludge [1].
Source: [2]
Genome Structure
N. moscoviensis has a circular chromosome [3], and its genome is 4.59 Mb in length. Its G+C content is 62% and it codes for approximately 4500 genes [4]. N. moscoviensis is hard to isolate in culture, and therefore research on its genome structure has been slow [2]. Its genome has been sequenced. However due to a lack molecular functional markers, detection methods have only recently been developed. Currently, the best available method is to track the gene encoding beta subunit of nitrite oxidoreductase or nxrB [5].
Figure 1:Newly designed PCR primers targeting the nxrB genes of Nitrospira [6]:
| Primer | Sequence (5’ - 3’) | Target Site | Amplicon size (bp) | Specificity |
|---|---|---|---|---|
| nxrB14f | ATA ACT GGC AAC TGG GAC GG | 14-33 | 1245 | N. moscoviensis, N. defluvii, and N. bockiana |
| nxrB 1239r | TGT AGA TCG GCT CTT CGA CC | 1239-1258 | ||
| nxrB 19f | TGG CAA CTG GGA CGG AAG ATG | 19-39 | All Nitrospira lineages | |
| nxrB 1237r | GTA GAT CGG CTC TTC GAC CTG | 1237-1257 | 1239 | |
| nxrB 169f | TAC ATG TGG TGG AAC A | 169-184 | All Nitrospira lineages | |
| nxrB 638r | CGG TTC TGG TCR ATC A | 638-653 | 485 |
In 2010, the first complete genome of a Nitrospira species, Nitrospira defluvii, was sequenced. Nitrospira defluvii was sequenced first because it was easier to culture and it has a smaller genome[4].
Cell Structure, Metabolism and Life Cycle
N. moscoviensis is an exclusively terrestrial, Gram negative, and non-motile bacteria. Unique features of species structure are an enlarged periplasmic space, a lack of cytoplasmic membrane, and carboxysomes. It also excretes nondescript polymers to form biofilms[1][7]. Nondescript polymers lack distinctive features; in other words, it can be thought of as a "general slime".
When it is exposed to 7.5 mN nitrite within a mineral medium, it takes 12 hrs to double in population size. N. moscoviensis undergoes binary fission to replicate[1].
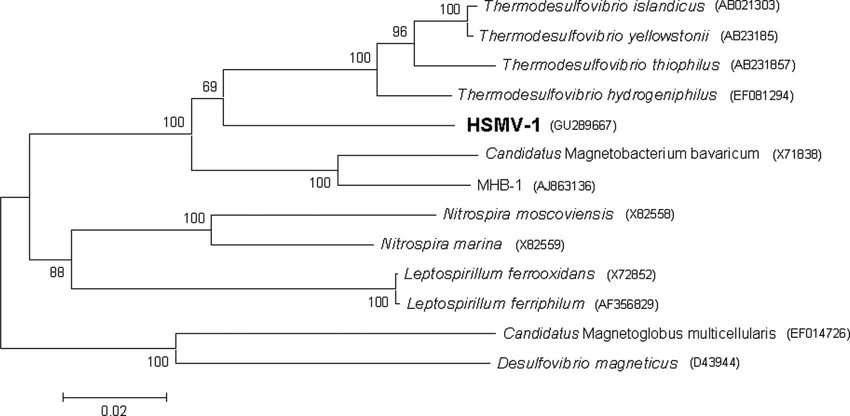
Figure 2: Analysis of 16S rRNA places N. moscoviensis close to Nitrospira marina on a phylogenetic tree [1].
Source: [[3]]
N. moscoviensis has high metabolic versatility. It is a chemolithoautotroph and a facultative aerobe. Under oxygenic conditions, it uses nitrite as its sole energy source and carbon dioxide as its sole carbon source. Organic matter does not induce mixotrophic or heterotrophic growth [1].
However, under anoxic conditions, N. moscoviensis uses hydrogen as an electron donor and nitrate as an electron acceptor, but the mechanism remains unclear. A solution containing soluble formate dehydrogenase allows it to use formate (CHOO-) as an electron acceptor as well. Furthermore, N. moscoviensis possesses the enzyme urease, which allows it to cleave urea into ammonia and carbon dioxide [7].
Ecology
N. moscoviensis is involved the second phase of the nitrogen cycle, in which it converts nitrite into nitrate. It also has the ability to cleave urea, this makes it incredibly useful in wastewater treatment plants. In these plants, N. moscoviensis is usually present in the activated sludge. In the presence of ammonia-oxidizing bacteria, it forms a symbiotic relationship known as reciprocal feeding. N. moscoviensis cleaves urea to form ammonia and carbon dioxide, and in return, the ammonia-oxidizing bacteria convert ammonia into nitrite, the preferred energy source of N. moscoviensis [7].
References
[1] Ehrich, S., Behrens, D., Lebedeva, E., Ludwig, W., & Bock, E. (1995). A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Archives of Microbiology,164(1), 16-23. [doi:https://doi.org/10.1007/BF02568729]
[2] Daims, H., Nielsen, J. L., & Schleifer, P. H. (2001). In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Applied and Environmental Microbiology,67(11), 5273-5284. [4]
[3] KEGG Genome: Nitrospira moscoviensis. (n.d.). Retrieved from [5]
[4] Nitrospira moscoviensis (ID 39992). (n.d.). Retrieved from [Organism:noexp]
[5] Lücker, S., Wagner, M., Maixner, F., Pelletier, E., Koch, H., Vacherie, B., . . . Daims, H. (2010). A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. PNAS,107(30), 13479-13484. [6]
[6] Maixner, F., Berry, D., Rattei, T., Koch, H., Lücker, S., Nowka, B., . . . Daims, H. (2013). NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite‐oxidizing Nitrospira. Environmental Microbiology,16(10), 3055-3071. [7]
[7] Koch, H., Lücker, S., Albertsen, M., Kitzinger, K., Herbold, C., Spieck, E., . . . Daims, H. (2015). Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. PNAS,112(36), 11371-11376. [doi:https://doi.org/10.1073/pnas.1506533112]
Author
Page authored by Casey DeCaro and Ashley DeRidder, students of Prof. Jay Lennon at Indiana University.