Photorhabdus luminescens
Classification
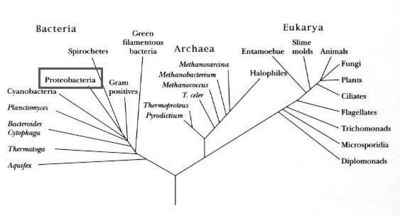
Domain: Bacteria, Phylum: Proteobacteria, Class: Gammaproteobacteria, Order: Enterobacteriales, Family: Enterobacteriaceae, Genus: Photorhabdus, Species: luminescens
Species
Photorhabdus luminescens
Description and Significance
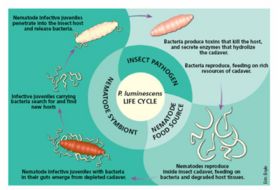
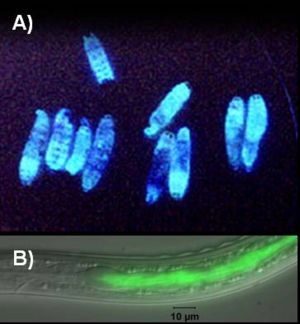
Photorhabdus luminescens also known as Xenorhabdus luminescens is a bioluminescent microbe (Figure 1). The role of bioluminescence in Photorhabdus luminescens is not well known.It might be used to warn other insects or mammals or to work as lure to attract insects for nematodes in which they live. The most important function of this microbe is the symbiotic relationship with soil entomopathogenic nematodes(family: Heterorhabditidae).They are also pathogenic to a wide range of insects (Figure 2).When the nematode infects an insect, P. luminescens is released into the blood stream of the insect and kills it by producing toxins. It also secretes enzymes that are responsible to break down the body of the infected insect and convert it into nutrients.These nutrients are later utilized by both nematode and bacteria. In this way, both organisms gain enough nutrients for further reproduction & replication. P. luminescens is the only organism found so far which exhibit dual phenotype (symbiotic within one insect, and pathogenic with another) (Figure 3).P. luminescens produces antibiotics and toxins to prevent invasion of the insect by bacterial or fungal competitors and it becomes visibly even in notocurnal environment due to the bioluminescence. There have been number of reported cases of human infection by Photorhabdus luminescens too.

Genome Structure
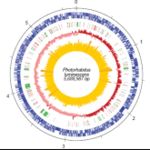
Photorhabdus luminescens genome sequence comprises of around 5,000 K base pairs and contains 5000 protein-coding genes (Duchaud et al.(2003)(Figure 4). The encoded genes consist of large arrays of adhesins, toxins, hemolysins, proteases and lipases. These proteins play vital role in killing competitors, host colonization, invasion and bioconversion of the insect cadaver.Thereby making this bacteria a very good model for the study of symbiosis and pathogensis.Genes that account for symbiotic relationships are also encoded on geneome sequences.Since the genome of P.luminescens is already sequenced (Duchaud et al. 2003)the next challenge is to identify genes involved in symbiosis which in future could be used to increase the production of the worms for controlling insects on plant. In on of the prototype experiments, Photorhabdus luminescens reduced Colorado potato beetles and sweet potato whitefly by 100 percent in lab conditions (Blackburn et al.(2005)). The potato beetle is notorious for developing resistance to insecticides, so scientists are seeking non-chemical controls as possible natural insecticides.
Cell Structure, Metabolism and Life Cycle
The life cycle of Photorhabdus luminescens is dependent on its symbiotic relationship with Heterorhabditis nematodes (Figure 3).Photorhabdus luminescens can be thought of as starting in the intestine of these nematodes. Once the nematode finds a suitable host insect the bacteria are released into the insect. While there they reproduce as do the nematodes. Once the infection stage is complete the bacteria find their way back into the intestine of the nematodes and the nematodes with their bacteria counterparts leave the old host in search of a new host to repeat the cycle. Photorhabdus luminescens exists in two distinct phases called phase I and phase II variants. The two variants have several differences in their cell expression. Phase I variants occur in infective-stage nematodes while Phase II variants occur after sustained growth in vitro. The phase I variant of Photorhabdus luminescens produces an extracellular protease, an extracellular lipase, antibiotic substances, and intracellular protein crystals while the phase II variant does not produce these substances. The phase I variant also has the remarkable trait of bioluminescence. The intracellular protein crystals are expressed as a result of the cipA and cipB genes which are present in the genome. The phase II variants do not express the cipA or cipB gene.Photorhabdus luminescens can be classified as an anaerobic organoheterotroph. The chemical processes responsible for metabolism occur in the absence of oxygen and organic carbon is used as the food source.
Ecology and Pathogenesis
The Photorhabdus luminescens bacterium is involved in a complex symbiotic relationship with certain entomopathogenic nematodes. But before the symbiosis can be understood the parasitic nature of the nematodes must be stated. These nematodes are parasitic for many types of insects, so much so that they are often used as a form of insect control across the world. Insects infected with these nematodes usually do not survive. But the nematode could never be lethal or proliferate on its own. It owes its continued existence to the symbiotic relationship it has with Photorhabdus luminescens which live in the intestine of these nematodes. Once inside the insect the nematodes systematically release Photorhabdus luminescens cells into the insect. Releasing as few as 10 cells into the insect is usually fatal. The process by which these bacteria avoid the insects natural defense mechanisms are currently unknown. The Photorhabdus luminescens cells inside the insect produce toxins and proteins which damage the host insect. One such toxin is the Mcf(makes caterpillars floppy) toxin (Figure 5). The Photorhabdus luminescens bacteria also are able to produce antibiotics which reduce competition between other bacteria. While inside the insect both the nematode and bacteria reproduce. After the infection is complete nematodes emerge once again living with Photorhabdus luminescens and the process can repeat with new insect hosts.
References
Blackburn, M. B. et al. The broadly insecticidal Photorhabdus luminescens toxin complex a (Tca): Activity against the Colorado potato bettle, Leptinotarsa decemlineata, and sweet potato fly Bemisia tabaci, Journal of Insect Science, 1-11 (2005)
Duchaud, E. et al. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nature Biotechnology 21, 1307-1313 (2003)
Liu, D. et al. Insect resistance conferred by 283-kDa Photorhabdus luminescens protein TcdA in Arabidopsis thaliana. Nature Biotechnology 21, 1222-1228 (2003)
Williamson et al. Sequence of a symbiont. Nature Biotechnology 21, 1294 - 1295 (2003)
Daborn, P. J., N. Waterfield, C. P. Silva, C. P. Au, S. Sharma, and R. H. ffrench-Constant. A single Photorhabdus gene, makes caterpillars floppy (mcf), allows Escherichia coli to persist within and kill insects. Proc. Natl. Acad. Sci. USA 99:10742-10747(2002)
Saux, M. F et al. Polyphasic classification of the genus Photorhabdus and proposal of new taxa: P. luminescens subsp. luminescens subsp. nov., P. luminescens subsp. akhurstii subsp. nov., P. luminescens subsp. laumondii subsp. nov., P. temperata sp. nov., P. temperata subsp. temperata subsp. nov. and P. asymbiotica sp. nov, International Journal of Systematic Bacteriology, 49, 1645-1656 (1999)
http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genomeprj&cmd=ShowDetailView&TermToSearch=9605
http://staff.bath.ac.uk/bssnw/photorhabdus_luminescens.htm
http://www.wormbook.org/chapters/www_genomesHbacteriophora/genomesHbacteriophora.html
http://www.genomenewsnetwork.org/articles/10_03/toxic_glow.shtml
http://www.ebi.ac.uk/2can/genomes/bacteria/Photorhabdus_luminescens.html
Author
Page authored by Ahsan Munir & Brian Charles Mcmillen ,student of Prof. Jay Lennon at Michigan State University.