Plasmavirus
Classification
Superkingdom: Viruses, Family: Plasmaviridae, Genus: Plasmavirus
Species
Plasmavirus L2
Description and Significance
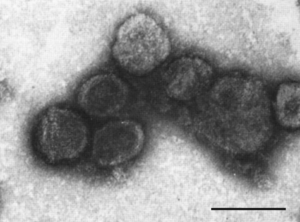
Plasmavirus L2, part of the Plasmaviridae family, is a bacteria-infecting virus with only one known host, being Acholeplasma laidlawii. 6 There is only one genus in the Plasmaviridae family, being Plasmavirus, and only one species Plasmavirus L2 (or Acholeplasma virus L2). 1 Little is known about the Plasmaviridae family and because it is poorly studied the diversity and biology of these viruses is largely unknown. The virion are enveloped, spherical to pleomorphic, with no head-tail structure ranging from diameters of 50-125 nm. The capsid is about 80 nm in diameter. 3 The absence of a regular capsid structure suggests plasmavirus virions consist of a condensed nucleoprotein bounded by a proteinaceous lipid vesicle. 3
Genome Structure
Plasmavirus L2 is a virion that is part of a family of bacteriophages, having an envelope, nucleoprotein complex, and a capsid. 5 They are between 50-125 nanometers in diameter, with an average of around 80 nm, and have a loose membrane surrounding them. They have a condensed, non-segmented genome that is a single molecule of circular, double-stranded, supercoiled DNA, being ~12000 bp in length. It has a dsDNA genome of about 12 kbp, encoding for at least 14 ORF (open reading frames) . 5
The L2 genome that has been sequenced and analyzed is 11,965 bp long with 15 open reading frames that are clustered into four groups by noncoding intergenic regions, implying that the gene expression involves transcription of genes in a cluster into polycistronic mRNA 6 (process where multiple separate proteins are being coded within one molecule) and translation of said genes via reinitiation or coupling.
Cell Structure, Metabolism and Life Cycle
DNA replication is bidirectional from two origins and is dependent on the host DNA replisome. The genus Acholeplasma virus L2 is extremely infectious to Acholeplasma laidlawii, with a G+C content of 32% which is similar to that of its host (31.8%). 8 At least three distinct virion forms are produced during infection (~75% of virions are 70–80 nm, ~20% are 80–90 nm, and ~5% are 110–120 nm). The three forms have the same protein composition, with differences in the relative amounts of each protein, but vary with respect to the number of encapsidated genome copies. 8 Virions are extremely heat-sensitive, relatively cold-stable and inactivated by nonionic detergents, ether and chloroform. The life cycle of Plasmavirus L2 consists of a cytoplasmic viral replication with DNA templated transcription. 6 The virus infects the host cell through fusion and exits the host cell by budding but may also become dormant within the host. Not enough is known about Plasmavirus L2 to identify its assembly or transmission site as of now.
Ecology and Pathogenesis
Plasmaviruses infect only one known species of Acholeplasma, which is a wall-less bacteria of the class Mollicutes, and release by budding through the cell membrane without causing host cell lysis. 9 The viral genome is then integrated into the host chromosome and transcribed by host polymerase and a productive infectious cycle begins before a lysogenic cycle (process of integration into the host chromosome) begins within the infected bacteria. After the initial infection by the viral genome the virus may become dormant within the host. 3 Although Acholeplasma laidlawii, a temperate bacteriophage, is the only current classified host of plasmavirus, related prophages reside in the genomes of different Acholeplasma species, where they are integrated into tRNA genes. Translational coupling or reinitiation may be involved in translation of the viral polycistronic mRNAs by the host translation machinery but not enough is known about the species to be sure. 8
References
1. U.S. National Library of Medicine. (2020). Taxonomy browser (plasmavirus). National Center for Biotechnology Information. Retrieved November 16, 2022, from https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=10473
2. Taxonomic information: Plasmavirus;inf. Organism: Plasmavirus;inf. (n.d.). Retrieved November 16, 2022, from http://www.prf.or.jp/cgi-bin/orgget.pl?org=Plasmavirus%3Binf
3. SIB Swiss Institute of Bioinformatics | Disclaimer. (n.d.). Plasmavirus. SIB Swiss Institute of Bioinformatics | Disclaimer. Retrieved November 16, 2022, from https://viralzone.expasy.org/575?outline=all_by_species
4. Plasmavirus. wikidoc. (2012). Retrieved November 16, 2022, from https://www.wikidoc.org/index.php/Plasmavirus
5. Maniloff, J., Kampo, G. J., & Dascher, C. C. (1994). Sequence analysis of a unique temperature phage: mycoplasma virus L2. Gene, 141(1), 1–8. https://doi.org/10.1016/0378-1119(94)90120-1Plasmavirus - mindat.org. (n.d.). Retrieved November 16, 2022, from https://www.mindat.org/taxon-10482913.html
6. Poddar, S. K., & Maniloff, J. (1987). Mapping of mycoplasma virus DNA replication origins and termini. Journal of virology, 61(6), 1909–1912. https://doi.org/10.1128/JVI.61.6.1909-1912.1987Acholeplasma laidlawii. VH DB. (n.d.). Retrieved November 16, 2022, from https://www.genome.jp/virushostdb/2148
7. Plasmavirus l2. VH DB. (n.d.). Retrieved November 16, 2022, from https://www.genome.jp/virushostdb/46014
8. Krupovic, M. (2018). ICTV Virus Taxonomy Profile: Plasmaviridae. Journal of General Virology, 99(5), 617–618. https://doi.org/10.1099/jgv.0.001060
9. Acholeplasma laidlawii. VH DB. (n.d.). Retrieved November 16, 2022, from https://www.genome.jp/virushostdb/2148
Author
Page authored by Nathaniel Asbell, student of Prof. Bradley Tolar at UNC Wilmington.