Pseudoalteromonas haloplanktis
A Microbial Biorealm page on the genus Pseudoalteromonas haloplanktis
Classification
Higher order taxa
Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Pseudoalteromonadaceae; Pseudoalteromonas
Species
P. agarivorans, P. aliena, P. antarctica, P. arctica, P. atlantica, P. aurantia, P. byunsanensis, P. carrageenovora, P. chazhmella, P. citrea, P. denitrificans, P. distincta, P. elyakovii, P. espejiana, P. flavipulchra, P. ganghwensis, P. gracilis, P. haloplanktis, P. issachenkonii, P. luteoviolacea, P. maricaloris, P. marina, P. mariniglutinosa, P. nigrifaciens, P. paragorgicola, P. peptidolyticam P. phenolica, P. piscicida, P. porphyrae, P. prydzensis, P. rubra, P. ruthenica, P. sagamiensis, P. spiralis, P. spongiae, P. telluritireducens, P. tetraodonis, P. translucida, P. tunicate, P. ulvae, P. undina, P. viridis, P. whanghaensis, P. sp.
|
NCBI: Taxonomy Genome: -P. haloplanktis TAC 125 chromosome 1 -P. haloplanktis TAC 125 chromosome II |
Description and significance
Pseudoalteromonas haloplanktis is a curved, rod shaped (0.2 to 1.5 by 1.8 to 3 µm), Antarctica psychrophile bacterium [1]. It was initially called Alteromonas tetraodonis, but has been reclassified as Alteromonas haloplanktis then to Pseudoalteromonas haloplanktis from the phylogenetic assays that was done by [1]. In addition, Pseudoalteromonas haloplanktis is a Gram negative bacterium that is non-spore-forming, not luminescent, and is motile by polar flagella [1]. This organism was isolated from the Antarctic coastal sea sample water and is found to grow only in marine environment at ~20ºC [3]. Researchers have sequenced some of its genome such as DNA ligase and cellulase to give an understanding of how its enzymatic proteins allow it to adapt at such low temperature [1].
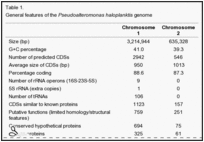
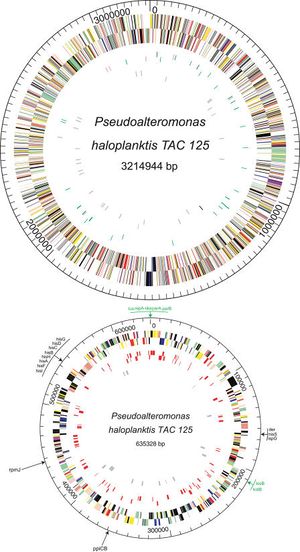
Genome structure
Pseudoalteromonas haloplanktis is an aerobe that has no fermentative metabolism [1]. They are rod-shaped bacteria that are classified as sub species: Pseudoalteromonas haloplanktis subsp. haloplanktis and Pseudoalteromonas haloplanktis subsp. tetraodonis [2]. Strain Pseudoalteromonas haloplanktis TAC 125 is completely sequenced and found to have 2 circular chromosomes, Chromosome I and Chromosome II [11]. Chromosome I has 41.0% G+C content with 2942 proteins while Chromosome II has 39.3% G+C rich with 546 proteins [9]. Genes which are encoded account for 87% of the genome [9]. An analysis of Pseudoalteromonas haloplanktis TAC125 genome sequence suggests that the increase in the degree of solubility (ie. turning the membrane into fluid)of oxygen at low temperature conditions is due to the bacterium replicating dioxygen scavenging cells so to destroy paths that lead to the creation of oxygen species [9]. The dioxygen cells receive safety and enable the opportunity to synthesize more lipids [9].
Cell structure and metabolism
The cell structure of the lipooligosaccharide (LOS) fraction isolated from the P. haloplanktis bacterium, consists of a linear-shaped inner core and no O-antigen side chain, which emphasizes the microorganism's ability to survive in cold conditions [3].P. haloplanktis consists of many cold-adapted enzymes and the majority of them have strong catalytic activities at lower temperatures with minimal activation energy needed to function[4]. P. haloplanktis is chemoautotroph that requires oxygen; it does not have fermentative metabolistic abilities [1]. The marine bacterium, Pseudoalteromonas haloplanktis TAC125, also has metabolic adaptability and has a good example of a psychrophilic microorganism [4]. The psychrophilic bacteria grow and survive in cold conditions that are usually below 5 °C; Because of these cold conditions the microorganisms have evolved through various intricate adaptations strategies that are focused on the cellular and extra-cellular structures of the bacteria [3]. The process of secreting proteins into these extra-cellular environments is very important so to supply quick and minimum amounts of energy needed to recover those recombinant proteins that were effected [8]. The significantly low temperature- adapted protein secretion system utilizes the psychrophilic α-amylase, which can be obtained from the P. haloplanktis bacterium [8]. Pseudoalteromonas haloplanktis DNA ligase has similar catalytic activity to other NAD+-dependent DNA ligases that are also low-temperature adaptation organisms [10]. The microorganism is seen to be controlled by the functions of its enzymes- the higher the reaction by the catalyzed protein, increases the metabolic flow and thus the cellular activities occur at low temperature conditions [6]. The need for lower temperatures of enzyme activity is the precise effect that offsets the relationship of the cold temperatures on biochemical reaction outcomes and what makes this gamma-proteobacterium so different in comparison to other bacteria found in water environments [8].
Pathology
Oysters are marine eukaryotic hosts for the bacterium, Pseudoalteromonas haloplanktis [7]. The pathologies in fish hosts does not occur often but the known toxic species of the bacteria is P. tetraodonis (Pseudoalteromonas haloplanktis tetraodonis), which is found in puffer fish (Fugu poecilonotus) [7]. The bacterial strain creates a potent substance called neurotoxin tetraodontoxin, which is responsible for severe intoxication and deaths reported in humans located in areas of Japan and other countries, through ingestion of insufficiently prepared puffer fish [7].
Application to Biotechnology
Pseudoalteromonas haloplanktis is seen as a valuable biotechnical microorganim because of the ability to grow fast and survive at low temperature conditions in the environment. Research of P. haloplanktis has begun to observe the effects of the bacterium as a new alternative expression host [5]. It has been shown that numerous heterologous enzymes can be produced through the soluble and active states of the bacterium and that P. haloplanktis contains a cold-adapted expression system for recombinant protein production [5].
References
Edited by Victoria Campbell, a student of Grace Lim-Fong
