Rhizobium bacteria nitrogen fixation
Introduction
Nitrogen is one of the most fundamental elements necessary for all life forms. Nitrogen is a major component of amino acids, the most basic building blocks of various proteins that sustain the proper functioning of a living organism; DNA, the fundamental biochemical unit of heredity that stores information in living organisms, also requires nitrogen to build up. Though nitrogen is highly abundant in the atmosphere in the form of dinitrogen gas, this molecular form of nitrogen is inert in that the triple-bonding between the two nitrogen atoms makes the molecule extremely stable at normal temperature and pressure. Given the contribution of nitrogen to sustaining life on Earth, natural biological pathways that convert nitrogen gas into bio-accessible forms are of great ecological and evolutionary significance.
Rhizobium bacteria represent one of the groups that perform the service of biological nitrogen fixation [1] The genus Rhizobium, commonly known as rhizobia, includes species of various gram-negative alphaproteobacteria and betaproteobacteria that inhabit the root nodules of leguminous plants [2]. Rhizobium was first discovered and named at the end of the 19th century when scientists began to notice that atmospheric nitrogen was assimilated into the root nodules of legumes: the german agricultural chemist Hermann Hellriegel first discovered that leguminous plants took in atmospheric nitrogen and turned it into ammonium; later, the Dutch microbiologist Beijerinck explored the mechanisms by which nitrogen is fixed through legume root-nodules and identified the bacteria responsible for this function, the rhizobia [3] . Today, the pathways through which rhizobia fix nitrogen and the genetic and ecological regulations that control the process have been thoroughly studied.
Rhizobia-legume Symbiosis: substances exchange
The most characteristic feature of a rhizobium is its symbiotic relationship with the host plant. Once infecting a host, the rhizobium elicits the formation of nodules in the host’s roots where the bacterium inhabits and fixes nitrogen; within the nodule, rhizobia are modified into bacteroids and compartmented into symbiosomes surrounded by symbiosome membranes. This symbiotic relationship between rhizobia and legumes is considered mutualistic since there are clear evolutionary advantages for both sides: while the bacteria provide the plant with a valuable nitrogen source, the plant in exchange feeds the rhizobium with sugar; this process has been understood on a molecular level. Within the root nodules, rhizobia reduce N2 into NH3 via the nitrogenase enzyme complex [4]. A great challenge of the enzyme nitrogenase is its strict requirement of an anaerobic condition. Rhizobia perform aerobic respiration; this suggests that special adaptations are needed to maintain the efficiency of nitrogen fixation within the cell. Several mechanisms protect the enzyme from oxygen: the inner cortex of the nodule serves as a diffusion barrier that limits the movement of oxygen into the rhizobium cell [5]; the cellular respiration of rhizobia within the nodule is enhanced by the high-oxygen-affinity terminal oxidase which leads to a greater rate of oxygen consumption; and finally, the leghaemoglobin, a specific protein that binds to oxygen, is produced in high concentrations by the roots colonized by rhizobia in order to buffer the oxygen concentration within root nodules [5] [4].
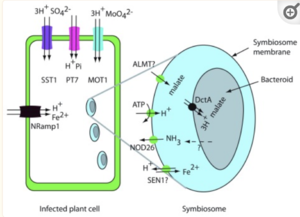
A functional nitrogenase carries out the energy-intensive process of nitrogen fixation that produces ammonia to be delivered to the host. In exchange, the host plant provides the bacteroids with carbon sources. In the legume cells infected by rhizobia-derived bacteroids within the nodule, the photosynthetic product of sucrose, which is cleaved by the reversible enzyme sucrose synthase, is the primary source of carbon for symbiotic metabolism going on inside nodules [6]. One study that used mutant peas with a low level of sucrose synthase activities showed that the enzyme is indispensable for the proper functioning of symbiotic nitrogen fixation; reduced levels of sucrose synthase decreased the leghemoglobin level by 80% which almost completely inhibited nitrogenase functioning [7]. Since rhizobia inhabit root cells that do not have access to sunlight, carbon must be transported within the host plant in order to deliver sucrose to the infected region. Research showed that the cells uninfected by bacteroids exhibited different metabolic activities from the infected cells where bacteroids perform nitrogen fixation: a series of transporter-regulated pathways translocate sucrose symplastically to the infected cells [8]. Within the infected region of root nodules, sucrose gets further processed. Under normal conditions, the cleaved sugar then subsequently feeds into glycolysis. However, in cells infected by rhizobia, the subsequent sugar metabolism exhibits alternative pathways. Evidence suggests that dicarboxylate, especially malate, is the final source of carbon that the host feeds to rhizobia; in root nodules, enzymes of the infected cells such as phosphoenolpyruvate carboxylase (PEPC) and malate dehydrogenase are greatly upregulated which divert carbon intermediates away from the glycolytic pathways and into malate synthesis [5] [9]. The malate then gets transported across the symbiosome membrane via specialized membrane dicarboxylate transporters and finally reaches the bacteroids; it is interesting that, besides carbon, the bacteroid also receives amino acids from the host via the symbiosome membrane as necessary building blocks of enzymes before nitrogen fixation begins [5]. Malate could be used as the sole source of energy in bacteroids; once the malate is provided, the bacteroid breaks it down through malic enzymes that decarboxylate malate into pyruvate [9]. Pyruvate is further processed into Acetyl-CoA, which feeds into the TCA cycle common to all forms of metabolism. This series of biochemical adaptations operated within both the host and the rhizobium provide a source of energy for the energy-intensive nitrogen fixation process.
Just as the host delivers carbon to its symbiotic partner, rhizobia need to transport the product of nitrogen fixation to the host to maintain the mutualistic relationship. The process of nitrogen fixation generates ammonia inside bacteroids; the ammonia will not be assimilated into organic forms but instead transported across the symbiosome membrane into the infected cell. Several mechanisms exist for this transmembrane ammonia transportation: since ammonia is a small weak acid molecule that exists in both charged and uncharged states, it can diffuse across the symbiosome membrane either through simple diffusion or via protein channels unspecific to ammonia; it can also be transported through active cation channels in the form of ammonium [5]. Ammonia is toxic when accumulated in great abundance, hence the cell needs to reprocess ammonia into alternative nitrogen forms to be transported within the host’s tissues; the form of nitrogen exported from the nodule to the rest of the host plant varies among species. For example, research suggested that purine synthesis is one of the pathways many legumes use to assimilate nitrogen fixed by rhizobia [10]. In tropical legumes such as soybeans and cowpeas, fixed nitrogen in forms of ammonia and ammonium is delivered completely to the purine pathway, which leads to the end product of ureides, the dominant form of stored nitrogen within the tissues of these legumes.
Development of the legume-rhizobia symbiosis
The depths of the soil host more biological activities than one might expect. It is a common phenomenon for plants or bacteria to communicate underground via diverse signaling mechanisms. For example, certain fungi are able to communicate with other individuals in response to environmental variations via electrical pulses. The development of a legume-rhizobia symbiotic relationship commonly relies on chemical signals sent out by the host. When a legume plant is in need of nitrogen sources, it begins to seek rhizobia around its roots; however, not all symbiotic relationships benefit the hosts as some parasitic microbes are pathogens that can harm their symbiotic partners. Hence, legumes have evolved the capacity to distinguish between beneficial and harmful bacteria through a series of chemical signaling pathways. Once triggered by nitrogen starvation, the host plant releases flavonoids, a group of polyhydroxy polyphenol, from the root in order to attract nearby rhizobia and cause them to migrate towards the root region via chemotaxis [11]. Besides attracting the right bacteria, flavonoids also induce the production of Nod factors in the targeted rhizobia, which are lipochitooligosaccharide signals that trigger the subsequent changes in the host’s root morphology. Once the LysM-type membrane receptors of the host recognize a nod factor, nodule formation and bacterial infection are induced that establishes the symbiotic relationship between the host and the selected rhizobium [12].
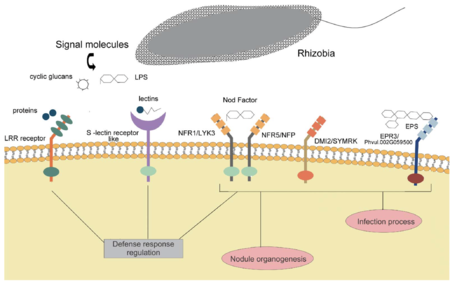
Specificity is a common characteristic of most rhizobia-legume symbiotic relationships. A specific host-symbiont relationship could result from long periods of coevolution during which both sides of symbiosis were adapted to be specific in functions. An individual species of rhizobia can colonize a narrow range of host species with the exception of Rhizobium sp. NGR234 that is able to nodulate a broad range of legumes [12]. The specificity is mediated by both the host and the bacteria. In general, legumes possess a diverse collection of flavonoids, but only a selected few are involved in symbiosis [13]. This acts as the first determinant of host-symbiont specificity in that the type of flavonoids excreted from the host’s root must have the capacity to induce nod factor production in the targeted rhizobia; for bacteria outside the range of symbiosis, this specific flavonoid will instead act as a phytoalexin that inhibits infection [13]. Rhizobia can similarly select ideal hosts by producing species-specific nod factors, which act as the second determinant of specificity. Though nod factors share the basic structural similarities as lipochitooligosaccharides, bacteria can modify the fatty acids attached to the chitooligosaccharide to produce nod factors with distinct structures [14]. Hosts thus are able to establish specific symbiotic relationships based on the recognition of nod factors by membrane receptors that bind to selected features. Though nod factors play the primary role in the development of symbiosis, cell wall components and secondary metabolites produced by rhizobia can also help regulate host specificity. For example, abundant research highlights the significance of exopolysaccharides in stabilizing the rhizobia-legume symbiosis; similar to nod factors, different bacteria also produce distinct exopolysaccharides that are regularly used by hosts to recognize specific rhizobia [15]. Other surface molecules such as lipopolysaccharides, cyclic glucans, and capsular polysaccharides are similarly evolved in either the recognition of symbiont or the suppression of the host’s immune defense upon infection [13].
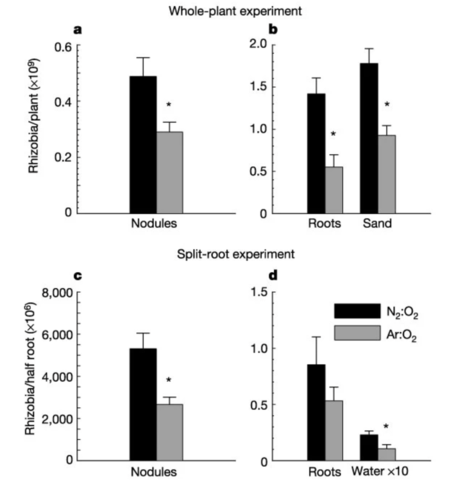
An interesting factor that plays a regulatory and stabilizing role in the rhizobia-legume symbiosis is the sanctioning mechanism, the rewarding of cooperation or the punishment of the non-cooperative ones. To maintain a mutualistic relationship, it is important for both sides to ensure that the benefit outweighs the costs associated with symbiotic nutrient exchanges. A reasonable assumption is that cheating behaviors might give one side extra advantages in a symbiotic relationship at the expense of its symbiotic partners; however, legumes have evolved host sanctions to keep cheating behaviors at bay. Commonly, a legume plant is simultaneously infected with several species of rhizobia; this implies that if the host does not allocate resources differentially to bacteria with various nitrogen-fixing abilities, natural selection will favor rhizobia that cheat, which would eventually break the mutualistic relationship. One study used experimental manipulations of air nitrogen levels to control the nitrogen-fixing capacity of rhizobia [16]. The results showed that, when a host was infected by multiple strains, bacteria that maintain a normal nitrogen-fixing level gained higher fitness due to greater resource allocation from the host and better reproductive success. The regulation of oxygen concentration within nodules seems to be an important mechanism through which the host plant controls the growth of symbiotic rhizobia [16]. In nodules colonized by non-nitrogen fixing bacteria, researchers reported both a decrease in interior oxygen content and a decrease in oxygen permeability through membranes. The sanctioning mechanism is likely to have evolutionary significance regarding the establishment and maintenance of the rhizobia-legume symbiosis [17].
Genetic regulations of nitrogen fixation and symbiosis
Many aspects of the rhizobial nitrogen fixation and the development of symbiosis with legumes are well understood on a genetic basis. In this section, I list a couple of important genes that are documented extensively throughout previous research.
The capacity of nitrogen fixation in rhizobia is granted by a range of nitrogen fixation genes; the nif family is a well-recognized collection of genes essential to nitrogen fixation. Nif genes encode the nitrogenase complex as well as regulatory proteins associated with the key enzyme nitrogenase; these genes are also found to regulate nitrogen fixation in other non-symbiotic bacteria such as cyanobacteria [5] [18]. Within the nif family, more than 10 individual genes have been identified to encode specific products: for instance, the nifH and nifD encode protein subunits of dinitrogenase reductase and dinitrogenase, two principal components of the nitrogenase complex; cofactors and regulators were encoded by genes such as nifA and nifBEN [18]. Nif genes provide regulations of nitrogen fixation with multiple levels: while nifA was demonstrated to be a positive regulator, nifL encodes a repressor [5]. Besides nif genes involved in the direct regulation of the nitrogenase complex, the fix family is another major class of genes essential to nitrogen fixation in rhizobia. The fix genes do not encode the structural components of nitrogen fixation, but they play key regulatory roles by interacting with nif genes. For example, fixL, fixK, and fixJ genes encode proteins that are involved in the sensing of low-oxygen signals (an anoxic environment is a prerequisite for nitrogen fixation), which then induce the expression of other nitrogen fixation genes [19]. There has been an established understanding that nif and fix genes works together in a cascade manner during the regulation of nitrogen fixation. In rhizobium, fixL and fixJ activate the nifA gene; the transmembrane protein encoded by fixL senses environmental signals for nitrogen fixation and transduces it to fixJ, which then activates nifA. The product of the nifA is an activator that induces the expressions of other nif and fix operons including nifHDK and fixABCX [20].
The genetic regulations of nodulation, the rhizobia-induced formation of nodules at the host’s roots, involve mainly the nod genes. As mentioned above, the symbiotic relationship between the host legume and rhizobia gets established when the flavonoids released by the host are recognized by the bacteria; the nodD gene encodes the critical transcriptional activator protein that functions as the receptor of flavonoids [21]. When bound to a flavonoid compound, the product of nodD, the NodD protein, goes through a subsequent conformational change; then the protein binds to the nod box, a conserved DNA sequence upstream of the inducible nod genes [22]. The following expression of the downstream nod genes produces various Nod factors, the lipooligosaccharides signals that rhizobia send to the host plant. When the signal is detected by a host plant, the plant goes through a subsequent series of morphological and physiological transformations: the curling of root hairs, bacterial infection into the root cortex through infection thread, and the formation of root nodules. These processes are regulated by nodulins, plant proteins essential for symbiotic nitrogen fixation encoded by nodulation genes [23].
Conclusion
Rhizobia is a special group of gram-negative proteobacteria that forms symbiotic relationships with legume plants. When a rhizobium colonizes and invades the root of a host, it stimulates nodule formations. Many previous studies focused on this symbiotic relationship since it represents one of the few natural pathways of nitrogen fixation, an energetically costly yet critical process of generating bioaccessible nitrogen essential for the functioning of all life forms. Rhizobia interact with potential hosts through chemical signaling, which gives rise to the subsequent steps of chemotaxis, special gene expression, bacterial infection of the host’s roots, and the morphological and physiological transformation of the host. These processes lead to the establishment of a mutualistic symbiotic relationship between rhizobia and legumes, which is characterized by necessary exchanges of nutrients and species-specific relationships. Like other non-symbiotic nitrogen fixers such as cyanobacteria, rhizobia rely on the nitrogenase enzyme complex to fix atmospheric nitrogen into ammonia. The proper functioning of this crucial enzyme is guaranteed by the nodule environment where anaerobic conditions are maintained and stable carbon sources are provided to the cell. The ammonia generated by rhizobia is not used by the bacteria but instead transported across membranes and supplied to the host cells.
In the age of climate change, the predicted trend of global warming and changes in weather patterns present potential threats as regards to agricultural productivity. The industrial production of crops relies on artificial fertilizers containing necessary nutrients including nitrogen; the industrial alternative has long been using the Haber-Bosch process to synthesize bioaccessible nitrogen, during which great energy is applied to nitrogen gas to overcome the energy barrier in order to forward the chemical reaction. However, there are environmental concerns associated with the use of nitrogen-containing industrial fertilizers [24]. Nitrogen fertilizers can be oxidized to form nitrous oxide, a potent greenhouse gas with a long residence time; the inorganic nitrogen can also be carried out by surface runoff and then feed into aquatic ecosystems, causing nitrogen pollution. The efficiency of nitrogen fixation granted by the rhizobia-legume symbiosis provides a great opportunity for reducing fertilizer use. It has been suggested that rhizobia can serve as a plant growth promoter by enhancing nutrient assimilations and improving plant defense against pests and pathogens [25]. Due to the diversity of the rhizobia, variants exhibit the potential to enhance the performance of leguminous crops in a variety of pH, temperature, and salinity conditions [25]. More future research is still required to explore the role of rhizobial symbiotic-nitrogen fixation in enhancing agricultural productivity, but it represents an opportunity to combat and adapt to climate change via nature-based solutions.
References
- ↑ Kennedy, P., Leonforte, A., & Butsch, M. (2015). Plant Breeding for biological nitrogen fixation: A Review. Biological Nitrogen Fixation, 1071–1076. https://doi.org/10.1002/9781119053095.ch106
- ↑ Willems, Anne. “The Taxonomy of Rhizobia: An Overview.” Plant and Soil 287, no. 1 (2006): 3–14. https://doi.org/10.1007/s11104-006-9058-7.
- ↑ https://bio.libretexts.org/Bookshelves/Microbiology/Book%3A_Microbiology_(Boundless)/5%3A_Microbial_Metabolism/5.15%3A_Nitrogen_Fixation/5.15B%3A_Early_Discoveries_in_Nitrogen_Fixation#:~:text=Martinus%20Willem%20Beijerinck%20
- ↑ 4.0 4.1 Udvardi, M., & Poole, P. S. (2013). Transport and metabolism in legume-rhizobia symbioses. Annual Review of Plant Biology, 64(1), 781–805. https://doi.org/10.1146/annurev-arplant-050312-120235
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 Lindström, Kristina, and Seyed Abdollah Mousavi. “Effectiveness of Nitrogen Fixation in Rhizobia.” Microbial Biotechnology 13, no. 5 (December 4, 2019): 1314–35. https://doi.org/10.1111/1751-7915.13517.
- ↑ Roy, Rahul, Anke Reinders, John M Ward, and Tami R McDonald. “Understanding Transport Processes in Lichen, Azolla–Cyanobacteria, Ectomycorrhiza, Endomycorrhiza, and Rhizobia–Legume Symbiotic Interactions.” F1000Research 9 (January 23, 2020): F1000 Faculty Rev-39. https://doi.org/10.12688/f1000research.19740.1.
- ↑ Gordon, null, null Minchin, null James, and null Komina. “Sucrose Synthase in Legume Nodules Is Essential for Nitrogen Fixation.” Plant Physiology 120, no. 3 (July 1999): 867–78. https://doi.org/10.1104/pp.120.3.867.
- ↑ Peiter, Edgar, and Sven Schubert. “Sugar Uptake and Proton Release by Protoplasts from the Infected Zone of Vicia Faba L. Nodules: Evidence against Apoplastic Sugar Supply of Infected Cells.” Journal of Experimental Botany 54, no. 388 (July 2003): 1691–1700. https://doi.org/10.1093/jxb/erg191.
- ↑ 9.0 9.1 Booth, Nicholas J., Penelope M. C. Smith, Sunita A. Ramesh, and David A. Day. “Malate Transport and Metabolism in Nitrogen-Fixing Legume Nodules.” Molecules (Basel, Switzerland) 26, no. 22 (November 15, 2021): 6876. https://doi.org/10.3390/molecules26226876.
- ↑ Smith, P. M., and C. A. Atkins. “Purine Biosynthesis. Big in Cell Division, Even Bigger in Nitrogen Assimilation.” Plant Physiology 128, no. 3 (March 2002): 793–802.
- ↑ Oldroyd, Giles E. D. “Speak, Friend, and Enter: Signalling Systems That Promote Beneficial Symbiotic Associations in Plants.” Nature Reviews Microbiology 11, no. 4 (April 2013): 252–63. https://doi.org/10.1038/nrmicro2990.
- ↑ 12.0 12.1 12.2 Via, Virginia Dalla, María Eugenia Zanetti, and Flavio Blanco. “How Legumes Recognize Rhizobia.” Plant Signaling & Behavior 11, no. 2 (December 4, 2015): e1120396. https://doi.org/10.1080/15592324.2015.1120396.
- ↑ 13.0 13.1 13.2 Walker, Liam, Beatriz Lagunas, and Miriam L. Gifford. “Determinants of Host Range Specificity in Legume-Rhizobia Symbiosis.” Frontiers in Microbiology 11 (November 27, 2020): 585749. https://doi.org/10.3389/fmicb.2020.585749.
- ↑ Long, S R. “Rhizobium Symbiosis: Nod Factors in Perspective.” The Plant Cell 8, no. 10 (October 1996): 1885–98.
- ↑ Acosta-Jurado, Sebastián, Francisco Fuentes-Romero, Jose-Enrique Ruiz-Sainz, Monika Janczarek, and José-María Vinardell. “Rhizobial Exopolysaccharides: Genetic Regulation of Their Synthesis and Relevance in Symbiosis with Legumes.” International Journal of Molecular Sciences 22, no. 12 (January 2021): 6233. https://doi.org/10.3390/ijms22126233.
- ↑ 16.0 16.1 16.2 Kiers, E. Toby, Robert A. Rousseau, Stuart A. West, and R. Ford Denison. “Host Sanctions and the Legume–Rhizobium Mutualism.” Nature 425, no. 6953 (September 2003): 78–81. https://doi.org/10.1038/nature01931.
- ↑ Akçay, Erol, and Ellen L. Simms. “Negotiation, Sanctions, and Context Dependency in the Legume-Rhizobium Mutualism.” The American Naturalist 178, no. 1 (2011): 1–14. https://doi.org/10.1086/659997.
- ↑ 18.0 18.1 https://www-sciencedirect-com.libproxy.kenyon.edu/topics/biochemistry-genetics-and-molecular-biology/nif-gene
- ↑ Fischer, H M. “Genetic Regulation of Nitrogen Fixation in Rhizobia.” Microbiological Reviews 58, no. 3 (September 1994): 352–86.
- ↑ David, Michel, Marie-Line Daveran, Jacques Batut, Annie Dedieu, Odile Domergue, Jyotsna Ghai, Cecilia Hertig, Pierre Boistard, and Daniel Kahn. “Cascade Regulation of Nif Gene Expression in Rhizobium Meliloti.” Cell 54, no. 5 (1988): 671–83. https://doi.org/10.1016/S0092-8674(88)80012-6.
- ↑ “Regulation of Nodulation Gene Expression by NodD in Rhizobia. - PMC.” Accessed April 18, 2022. https://www-ncbi-nlm-nih-gov.libproxy.kenyon.edu/pmc/articles/PMC206349/.
- ↑ Göttfert, Michael. “Regulation and Function of Rhizobial Nodulation Genes.” FEMS Microbiology Reviews 10, no. 1–2 (1993): 39–63. https://doi.org/10.1111/j.1574-6968.1993.tb05863.x.
- ↑ “Regulators and Regulation of Legume Root Nodule Development | Plant Physiology | Oxford Academic.” Accessed April 18, 2022. https://academic-oup-com.libproxy.kenyon.edu/plphys/article/124/2/531/6098804.
- ↑ https://www.soilassociation.org/causes-campaigns/fixing-nitrogen-the-challenge-for-climate-nature-and-health/the-impacts-of-nitrogen-pollution/#:~:text=When%20used%20in%20excess%2C%20nitrogen,more%20potent%20than%20carbon%20dioxide
- ↑ 25.0 25.1 Mabrouk, Yassine, Imen Hemissi, Issam Ben Salem, Sonia Mejri, Mouldi Saidi, and Omrane Belhadj. Potential of Rhizobia in Improving Nitrogen Fixation and Yields of Legumes. Symbiosis. IntechOpen, 2018. https://doi.org/10.5772/intechopen.73495.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2022, Kenyon College
