Rhodococcus pyridinivorans
Classification
Kingdom: Bacteria
Phylum: Actinobacteria
Order: Actinomycetales
Suborder: Corynebacterineae
Family: Nocardiaceae
Genus: Rhodococcus
Species
Rhodococcus pyridinivorans (also named as pyridinovorans)
NCBI: Taxonomy
Description and Importance
In 2000, a novel species named as Rhodococcus pyridinivorans was first cultured from industral wastewater in Korea. It is a gram positive, aerobic, rod shaped, non-motile, and non-sporulating mesophiles. The sepcies named "pyridinivorans" (pyridine-devouring) was derived from its capability to degrade a pyridine (10).
Although the genus, Rhodococcus, implies the coccus shape of the organisms, the bacteria surprisingly show extensive polymorphism (Fig. 1.) (6). At the early stage of its growth, a Rhodococcus pyridinivorans bacterium is rod-shaped and filamentous, but it grows and breaks into a short rod or coccus (10).
R.pyridinivorans (stain PDB9T) has been characterized based on phylogenetic and chemotaxonomic descriptors such as G+C content, 16S rDNA sequences, and DNA-DNA relatedness. Its DNA G+C content (66 mol%) indicated that the organism falls within the genus Rhodococcus. The 16S rDNA analysis revealed 99% nucleotide similarity to that of the type strain of R.rhodochrous (Fig. 2) (10).
The fatty acid biography of the strain PDB9T is also similar to the type strains of Rhodococcus species, but the amounts of C16:0, C18:1 cis9, 10-methyl-C18:0 show some variations. Furthermore, its ability to degrade pyridine and the levels of DNA-DNA relatedness (Table 1) confers its distinctiveness. Thus, based on these differences, strain PDB9T was placed into a new species (10).
Recently, several other strains of R. pyridinivorans species were discovered (strain AK 37 and PA). These strains are not limited to pyridine and also have the capability to transform toxic, aromatic compounds. Strain AK 37, isolated from an oil-polluted soil in Hungary, degrades BTEX, an acronym standing for benzene, toluene, ethylbenzene, and o-xylenes (1, 7), and strain PA has the capability to degrade benzothialzole (8). The strains of this species exhibit a wide range of capabilities in degrading organic compounds. The major ecological importance of R. pyridinivorans derives from their roles in bioremediation.
Genome Structure
Among the three strains, the whole genome sequence of the strain AK35 has only been sequenced in order to better understanding benzothalzole (BT) degradation. Its genome is 5.2 Mb with 67.8% G+C content, containing 4,822 protein coding genes, three encode rRNA : one for 16S rRNA and two for 23S rRNA (GenBank database accession number AHBW00000000.1) (1).
For the strain PDB9T, its 16S rDNA was only sequenced for the classification. It is composed of 1477 nucleotides (GenBank accession number NR_025033.1), which showed 99% similarity to R.rhodochrous DSM 43241T (10). For the strain PA, its 16S rRna sequence (1,384 bases, GenBank accession number AJ457068.1) was analysed and compared to the 16S rDNA of the strain PDB9T, showing 100% match over the 1,384 bases of the strain PA (8).
Cell Structure and Metabolism
Unlike its genome structure, the cell structure of the strain PDB9T is well analyzed for its taxonomic position. The major components of its cell wall include meso-diaminopimelic acid, mycolic acids (36-46 carbon atoms), and arabinose and galactose as major carbohydrates. Glycosyl is present as the acyl type in the beta-1,4-N acetylmuramic acid. These distinctive chemotaxonomic markers have attributed to the differential of the strain PDB9T from other type strains of R. pyridinivorans. As it is stated above, the strain PDB9T is able to degrade pyridine, however, the exact mechanism of pyridine biodegradation has not been studied well (10).
The genome sequence of R.pyridinivorans strain AK37 has identified several key enzymes involved in the six pathways of monocyclic aromatic compound biodegradation: protocatechuate 3,4-dioxygenase, benzoate 1,2-dioxygenase, 3-ketosteroid-9α-hydroxylase, 3-ketosteroid-σ-dehydrogenase, and so on. Some of these enzymes seem to be widely distributed among aromatic-compound degrading species. THe pathway of aromatic compound degradation by strain AK 37 has not been described (1).
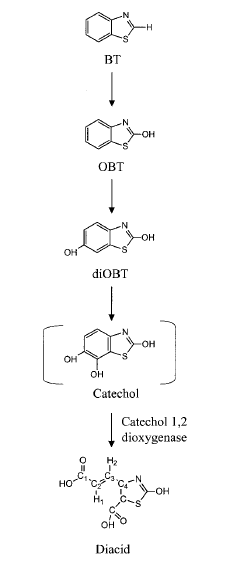
The cell structure of the strain PA has not been reported. Only has the pathway for BT biodegradation been proposed based on the identification of intermediate molecules (Fig. 3). A benzothiazole (BT) is first hydroxylated by a monooxygenase, producing 2-hydroxybenzothiazole (OBT). OBT is also hydroxylated by the same enzyme into diOBT. A final product of the proposed pathway, dicarboxylic acid, suggests the involvement of a catechol 1,2-dixoygenase in cleavage of the benzene ring (1,6C). Most parts of the BT degradation pathways are still unknown, and many uncertainties are currently being studied (8).
Ecology and Bioremediation
R. pyridinivorans species are commonly present in contaminated soil or water with a variety of xenobiotics, specifically toxic aromatic compounds(10). These compounds are produced by both anthropogenic and natural activities and accumulate in the environment (9). Many these aromatic compounds, including pyridine, are toxic with implications for public and ecosystem health.
Pyridine is a colourless, water-soluble compound with nauseating odor. It is widely used as a solvent in analytical chemistry and industry, and is an intermediate in the synthesis of herbicides, bactericides, and pharmaceuticals. It affects the nervous system and gastrointestinal tract of humans, causing headache, dizziness, nausea, and a lack of coordination. It also has acute effects on aquatic organisms such as fish (5). Benzothiazoles (BT) is released into the environment as a result of the applications as anti-algal agents, herbicides, fungicides, and slimicides in the paper industry. It is a recalcitrant molecule that is potentially carcinogenic and mutagenic, causing kidney and liver failure, central nervous system depression, dermatitis, and so on (2, 4). Finally, BTEX, derived from crude oil and petroleum products, poses a great public health and environmental concern due to its water solubility and potential carcinogenicity (7).
These xenobiotic and organic compounds are more efficiently degraded by R. pyridinivorans bacteria than in the nature. Thus, their biodegrading capabilities of the toxic compounds contribute greatly to bioremediation. This species has not been reported to being pathogenic.
References
1.Balazs Kriszt, et al. 2012. De Novo Genome Project for the Aromatic Degrader Rhodococcus pyridinivorans Strain AK37. J. Bacteriol. 194(5):1247-1248.
2.De Wever, H. & Verachtert, H. 1997. Biodegradation and Toxicity of Benzothiazoles. Elsevier Science Ltd. 31(11):2673-2684.
3.Gaja, M., A. and Knapp, J., S. 1997. The Microbial Degradation of Benzothiazoles. Journal of Applied Microbiology. 83:327-334.
4.Gary Ginsberg, Brian Toal & Tara Kurland. 2011. Bezothialzole Toxicity Assessment in Support of Synthetic Turf Field Human Health Risk Assessment. Journal of Toxicology and Environmental Health. 74(17):1175-1183.
5.Jori, A., et al. 1983. Ecotoxicological Profile of Pyridine. Ecotoxicology and Environmental Safety. 7:251-275.
6.Larkin, M., J., Kulakov, L., A., and Allen, C., C., R. 2010. Rhodococcus. School of Biological Sciences and The QUESTOR Centre, The Queen’s University of Belfast, Belfast, Northern Ireland, UK. 2:1840-1852.
7.Margesin, R., Walder, G., Schinner, F. 2003. Bioremediation Assessment of a BTEX-Contaminated Soil. Acta Biotechnol. 23(1):29-36.
8.Nicolas Haroune, et al. 2002. Bezothiazole Degradation by Rhodococcus pyridinovorans Strain PA: Evidence of a Catechol 1,2-Dioxygenases Activity. Appl. Environ. Microbial. 68(12):6114-6120.
9.Shelly Shinha, et al. 2009. Microbial Transformation of Xenobiotics for Environmental Bioremediation. African Journal of Biotechnology. 8(22):6016-6027.
10.Yoon,J.-H., et al. 2000. Rhodococcus pyridinivorans sp. nov., a pyridine-degrading bacterium. Int. J. Sys. Evol. Microbiol. 50:2173-2180.
Edited by student of William Mohn at the University of British Columbia.