Riftia pachyptila symbiont

Characteristics of the bacterial symbiont
The bacterial symbiont of R. pachyptila has not been successfully cultivated apart from its host, therefore standard phenotypic methods of classification have not been applicable. Using DNA-DNA hybridization, the degree of phylogenetic similarity among R. pachyptila bacterial symbionts from widely distanced vent sites has been determined. While the symbiont could not be cultivated from the host, purified R. pachyptila symbiont DNA standards were able to be obtained. This was because the symbiont DNA could be physically separated from R. pachyptila DNA because of its higher G+C content and possession of high-density internal sulfur globules. Standard DNAs obtained were hybridized against trophosome DNAs obtained from distantly separated R.pachyptila individuals, as well as other vestimentiferan genera, to investigate symbiont similarities. The bacterial symbionts from the three vent sites studied were all members of the same bacterial species. The association between the host and symbiont is highly specific. The likely reason for the lack of host differentiation is that there is not enough time for accumulation of mutations to occur because of the rapid extinction and reestablishment of populations given the short lifespan of hydrothermal vents. Therefore the lack of genetic diversification observed in the host should extend to the symbiont [1].
Characteristics of Riftia pachyptila
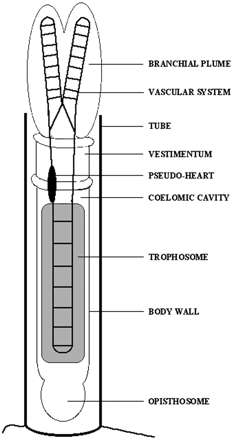
Riftia pachyptila is a giant tubeworm of typically one to two meters in length that inhabits the volcanic deep sea vents of the Pacific Ocean. A plume protrudes from the R. pachyptila protective tube and contacts the surrounding water. The plume has a large, highly vascularized surface which allows for the exchange of metabolites between R. pachyptila and the environment. Other tissues within the R. pachyptila tube include the vestimentum, which allows R. pachyptila to position itself in the tube, and the richly vascularized trophosome [2]. R. pachyptila does not have a digestive tract and must live in an obligate symbiosis with a sulfur-oxidizing chemoautotrophic bacterium. This mutualistic symbiont is localized in the R. pachyptila trophosome cells, which are densely colonized by the bacterium. [3]. The bacterium is estimated to represent as much as 35% of the total volume of the trophosome [4]. R. pachyptila larvae have a digestive tract which disappears during development, so it is likely the trophosome that is colonized with the bacterium each generation [5].
The circulatory system includes a pump located in the vestimentum region that promotes blood circulation in the entire body, including to the trophosome cells which bring nutrients to the bacterium. The plume is rich with blood, which can be visualized by the red color of the plume. The circulatory system mediates all metabolite exchanges between R. pachyptila and the surrounding water [6][7].R. pachyptila are adapted to their volcanic deep sea environment and use its composition, which include carbon, nitrogen, oxygen, and sulfur, in metabolic pathways that rely on the symbiotic relationship with the bacterium. The R. pachyptila hemoglobin is the transporter of both oxygen and sulfide to the bacterium which produce metabolic energy for both itself and R. pachyptila [8].
Host-Symbiont Interaction
Nutrition in R. pachyptila depends on the bacterial symbiont to fixate carbon dioxide using energy derived from the oxidation of reduced sulfur compounds. The bacterial symbiont oxidizes sulfide into sulfite by an electron transport system, which eventually results in the production of ATP that can be used by the symbiont for the assimilation of carbon.
Assimilation of Carbon
R.pachyptila absorb carbon dioxide produced by the surrounding hydrothermal vents using its brachial plume. Once absorbed, carbon dioxide can be used in many ways. Carbon dioxide can be transported by the circulatory system to the trophosome where bacteria are located. In addition, carboxylation in the plume results in malate, which can be transported immediately to the trophosome by blood circulation [9]. In the bacteria, the carbon dioxide from the plume provided either by the environment or as a result from the decarboxylation of the transported malate enters the Calvin-Benson cycle and serves as a precursor for different small organic metabolites. These metabolites, such as as ribulose-1,5-biphosphate and ribulose-5-phosphate, can be delivered to the different tissues of R. pachyptila for its own metabolism and ATP production [10].
Assimilation of Nitrogen
The bacterial symbiont has a high demand for nitrogen due to its large biomass and high growth rate. This is consistent with the high level of nitrate in the surrounding environment [11]. Ammonia resulting from the reduction of nitrate by the bacterial symbiont can be utilized by R. pachyptila as well as produce metabolites, such as amino acids and nucleotides, for the bacterial symbiont [12]. Ammonia along with carbon dioxide can also be used in the biosynthetic pyrimidine and arginine pathways.
R. pachyptila lack enzymes required for the de novo pyrimidine pathway as well as those required for the biosynthesis of polyamines, while the bacterial symbiont lacks enzymes required for the pyrimidine salvage pathway.
Pyrimidine metabolism
The de novo pathway, which utilizes carbon and nitrogen, and the salvage pathway, which utilizes nucleic acids, are the two metabolic pathways responsible for the production of pyrimidine nucleotides. Enzymes required in the pyrimidine de novo pathway are only present in the bacterial symbiont. R. pachyptila is unable to synthesize pyrimdine nucleotides through the de novo pathway and must rely on the salvage pathway. R. pachyptila contain all of the enzymes required for this pathway. R. pachyptila is completely dependent on the bacterial symbiont for the de novo biosynthesis of the pyrimidine nucleotides [12].
Arginine metabolism
Arginine carboxylase and ornithine decarboxylase have key roles in the synthesis of polyamines for the R. pachyptila cell tissue. Polyamines are involved in membrane stability and growth. R. pachyptila cannot utilize arginine metabolism because it lacks key enzymes and therefore must rely on the bacterial symbiont. Putrescine, the product of polyamine degradation, can serve as an alternative source of inorganic carbon and nitrogen for R. pachyptila [13].
Molecular Insights into the Symbiosis
Sulfide Acquisition
Sulfide and oxygen are both required for the bacterial symbiont to produce ATP. However, sulfide spontaneously reacts with oxygen to form sulfur compounds. This decreases the available free sulfide for the bacterial symbiont to oxidize. The bacterial symbiont must compete with oxygen for free sulfide and reside at the interface between oxic and anoxic zones so it can acquire oxygen but without prematurely oxidizing the free sulfide. The bacterial symbiont has adapted to this by residing with R. pachyptila [8].
Simultaneous acquisition of sulfide and oxygen occurs in R. pachyptila because of a biochemical adaptation. Sulfide typically interacts to inhibit oxygen-binding on hemoglobin,, however, the multi-hemoglobin complex synthesized by R. pachyptila reversibly binds sulfide independent of oxygen. Sulfide, primarily as hydrogen sulfide, and oxygen can then be transported by the R. pachyptila circulatory system to the trophosome for use by the bacterial symbiont [6][7].
Carbon Acquisition
The bacterial symbiont uses the Calvin-Benson cycle for carbon fixation and therefore requires carbon dioxide, which diffuses readily through biological membranes. While the majority of dissolved carbon in the sea is bicarbonate due to the higher pH of the sea (pH 8.0), the lower pH around hydrothermal vents (pH 6.0) generates higher concentrations of carbon dioxide and gives organisms that utilize the Calvin-Benson cycle an advantage. R. pachyptila must offset the proton-generating reaction of sulfur oxidation by bacterial symbionts to create the gradient required to intake carbon dioxide. R. pachyptila rely on H+-ATPases to export proton ions. This maintains R. pachyptila blood at an alkaline pH of 7.5 [8]. While the low pH of the surrounding hydrothermal vent water results in a greater carbon dioxide concentration, the alkaline pH of the R. pachyptila blood favors the conversion of carbon dioxide to bicarbonate, which establishes a carbon dioxide gradient across the R. pachyptila plume. This gradient, from higher external concentration of carbon dioxide to lower internal concentration of carbon dioxide, drives the diffusion of carbon dioxide into the R. pachyptila blood. Carbon dioxide is then transported by the blood to the trophosome for use by the bacterial symbiont in carbon fixation via the Calvin-Benson cycle [9].
Nitrogen Acquisition
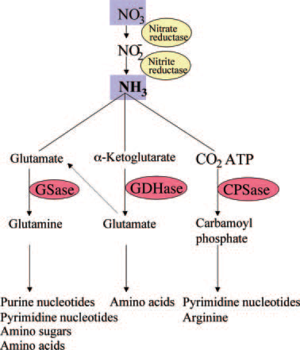
Nitrate reductase, the bacterial enzyme which mediates the reduction of nitrate to nitrite, has been detected in R. pachyptila, indicating a role for nitrate reduction in the assimilatory pathway by which nitrate is ultimately converted to ammonia for use in both R. pachyptila and symbiont biomass synthesis [14].
Glutamine synthetase (GSase) and glutamate dehydrogenase (GDHase) are the primary enzymes that mediate the assimilation of ammonia into amino acids. These enzymes have also been determined to be in R. pachyptila. While both R. pachyptila and the bacterial symbiont synthesize these enzymes, protein characterization showed that GSase measured in the trophosome was synthesized from the bacterial symbiont [15], implicating that the symbiont is responsible for inorganic nitrogen acquisition. Only the bacterial symbiont has all of the enzymes required for the de novo synthesis of pyrimidines, implying that R. pachyptila is dependent on the bacteria; symbiont for these nucleotides. While absent in the bacterial symbiont, the activities of three of the enzymes that mediate pyrimidine catabolism were detected in the R. pachyptila, suggesting that pyrimidine degradation may represent an internal source of carbon dioxide and ammonia for use in R. pachyptila biosynthesis [12]. The catabolic enzymes involved in the synthesis of polyamines from arginine appear to be present only in the bacterial symbiont [13]. Polyamines, which play important physiological roles in growth, membrane structure, and nucleic acid synthesis, and which represent a potential source of carbon and nitrogen upon degradation, may therefore be available to R. pachyptila only via the metabolism of the bacterial symbiont [13]. This emphasizes the extent to which the metabolism of R. pachyptila is affected by this symbiosis.
Ecological and Evolutionary Aspects
The evolutionary aspects between R. pachyptila and the bacterial symbiont depend heavily on the symbiont transmission strategy. Bacterial symbiont transmission can occur when R. pachyptila acquire the bacterium from a free-living population in the environment, or horizontally by transfer of bacteria between R. pachyptila sharing the same habitat.
R. pachyptila are completely dependent on the bacterial symbiont for nourishment, so environmental transmission would not seem favorable, since the hydrothermal vent environment is extreme. However, studies have shown that R. pachyptila acquires the bacterial symbiont de novo each generation from a population of free-living bacteria. These bacterial symbionts do not demonstrate cospeciation with their hosts. Analysis of 16S rRNA gene sequences showed that vestimentiferan tubeworms belonging to the genera Riftia, Oasisia, and Tevnia share a very similar symbiont phylotype [16]. The distribution of a single phylotype among these three vestimentiferan genera is evidence that these tubeworms acquire their bacterial symbionts from a free-living population of bacteria. PCR using R. pachyptila symbiont-specific primers did not detect bacterial 16 rRNA genes in DNA extracts from R. pachyptila eggs [5]. This suggests that the bacterial symbiont is not vertically transmitted. Since tubeworms during early development have a digestive tract, but mature tubeworms lack a digestive tract, bacterial symbiont cells in R. pachyptila eggs were not expected, since it is not until the tubeworms are mature that they become incapable of feeding on their own. Mature tubeworms rely on the bacteria-colonized trophosome, where carbon dioxide fixation takes place [5].
The detection of specific functional genes in the bacterial symbiont also suggests environmental transmission. Using PCR to detect and amplify a bacterial symbiont gene, it was discovered that a bacterial symbiont gene had high sequence similarity to the flagellin gene, fliC, which encodes the primary subunits of the bacterial flagellum. This shows that the bacterial symbiont has at least one of the genes required for flagellar synthesis. Flagellar motility would not be necessary if the bacterial symbiont was always associated with R. pachyptila, so this is further evidence that vertical transmission is unlikely. If the bacterial symbiont is environmentally transmitted, a flagellum could mediate adhesion R. pachyptila [17].
Recent Discoveries
While most studies on R. pachyptila and its bacterial symbiont have focused on their habitat in the basalt-hosted vents, recent studies have focused on R. pachyptila that reside in sediment-hosted vents. Differences in nitrogen metabolism were discovered in the bacterial symbiont in these sediment-hosted vent environments [18]. R. pachyptila in these environments utilize different sulfur compounds. Unlike R. pachyptila in basalt-hosted environments, R. pachyptila in sediment-hosted environments may assimilate reduced nitrogen. The surrounding environment heavily influences the way R. pachyptila and its bacterial symbiont interact [18].
Another recent study examined DNA sequences of R. pachyptila in basalt-hosted vents. R. pachyptila DNA sequence diversity was found to be extremely low [19]. This may be due to the harsh environment that the R. pachyptila inhabits. Volcanic and tectonic activities cause high rates of local extinction, while R. pachyptila recolonize only from a limited number of source populations. This harsh envrionment reduced genetic variance and contributes to the demographic instability of R. pachyptila [19].
References
[1] Edwards,D.B., Nelson, D.C. (1991) Edwards, D.B., Nelson, D.C. (1991) DNA-DNA solution hybridization studies of the bacterial symbionts of hydrothermal vent tube worms (Riftia pachyptila and Tevnia jerichonana). Applied and Environmental Microbiology 5, 1082–1088.
[2] Gaill, F. (1993) Aspects of life development at deep sea hydrothermal vents. FASEB Journal 7, 558–565.
[3]Hand, S.C. (1987) Trophosome ultrastructure and the characterization of isolated bacteriocytes from invertebrate-sulfur bacteria symbioses. Biol. Bull. 173, 260–276.
[4] Powell, M.A., Somero, G.N. (1986) Adaptations to sulfide by hydrothermal vent animals: sites and mechanisms of detoxification and metabolism. Biol. Bull. 171, 274-290.
[5] Nussbaumer, A.D., Fisher, C.R., Bright, M. (2006) Horizontal endosymbiont transmission in hydrothermal vent tubeworms. Nature 441, 345-348.
[6] Zal, F., Lallier, F.H., Wall, J.S., Vinogradov, S.N. & Toulmond, A. (1996) The multi-hemoglobin system of the hydrothermal vent tube worm Riftia pachyptila. I. Reexamination of the number and masses of its constituents. J. Biol. Chem. 271, 8869–8874.
[7] Zal, F., Lallier, F.H., Green, B.N., Vinogradov, S.N. & Toulmond, A. (1996) The multi-hemoglobin system of the hydrothermal vent tube worm Riftia pachyptila. II. Complete polypeptide chain composition investigated by maximum entropy analysis of mass spectra. J. Biol. Chem. 271, 8875–8881.
[8] Goffredi, S.K., Childress, J.J., Desaulniers, N.T. Lallier, F.J.(1997) Sulfide acquisition by the vent worm Riftia pachyptila appears to be via uptake of HS–, rather than H2S. J. Exp. Biol. 200, 2609–2616.
[9]Childress, J.J., Lee, R.W., Sanders, N.K., Felbeck,H., Oros,D.R., Toulmond, A., Desbruyeres, A., Kennicutt, M.C., Brooks, J.(1993)Inorganic carbon uptake in hydrothermal vent tubeworms facilitated by high environmental pCO2. Nature 362, 147–149.
[10] Robinson, J.J., Stein, J.L., Cavanaugh, C.M. (1998) Cloning and sequencing of a form II ribulose-1,5-biphosphate carboxylase/oxygenase from the bacterial symbiont of the hydrothermal vent tubeworm Riftia pachyptila. J. Bacteriol. 180, 1596–1599.
[11] Lee, R.W., Childress J.J. (1994) Assimilation of inorganic nitrogen by marine invertebrates and their chemoautotrophic and methanotrophic symbionts. Applied and Environmental Microbiology 60, 1852–1858.
[12] Minic, Z., Simon, V., Penverne, B., Gaill, F., and Herve, G. (2001) Contribution of the bacterial endosymbiont to the biosynthesis of pyrimidine nucleotides in the deep-sea tube worm Riftia pachyptila. J. Biol. Chem. 276, 23777–23784.
[13] Minic, Z., Herve, G. (2003) Arginine metabolism in the deep sea tube worm Riftia pachyptila and its bacterial endosymbiont. J. Biol. Chem. 278, 40527–40533.
[14] Girguis, P.R., Lee, R.W., Desaulniers, N., Childress, J.J., Pospesel, M., Felbeck, H., Zal, F.(2000) Fate of nitrate acquired by the tubeworm Riftia pachyptila. Applied and Environmental Microbiology 66, 2783–2790.
[15] Mimic, Z. Herve, G. (2004). Biochemical and enzymological aspects of the symbiosis between the deep-sea tubeworm Riftia pachyptila and its bacterial endosymbiont. Eur. J. Biochem 271, 3093-3102.
[16] Di Meo, C.A., Wilbur, A.E., Holben, W.E., Feldman, R.A., Vrijenhoek, R.C., Cary, S.C. (2000) Genetic variation among endosymbionts of widely distributed vestimentiferan tubeworms. Applied and Environmental Microbiology 66, 651–658.
[17] Millikan, D.S., Felbeck, H., Stein, J.L. (1999) Identification and characterization of a flagellin gene from the endosymbiont of the hydrothermal vent tubeworm Riftia pachyptila. Applied and Environmental Microbiology 65, 3129–3133.
[18] Robidart, J.C., Roque, A., Song, P., Girguis, P.(2011) Linking hydrothermal geochemistry to organismal physiology: physiological versatility in Riftia pachyptila from sedimented and basalt-hosted vents. PLoS One. 6: e21692.
[19] Coykendall, D.K., Johnson, S.B., Karl, S.A., Lutz, R.A., Vrijenhoek, R.C. (2011) Genetic diversity and demographic instability in Riftia pachyptila tubeworms from eastern pacific hydrothermal vents. BMC Evol. Biol. 11: 96.
Edited by [Crystal Leibrand], students of Grace Lim-Fong
This template is just a general guideline of how to design your site. You are not restricted to this format, so feel free to make changes to the headings and subheadings and to add or remove sections as appropriate.
