SARS-CoV: nsp7 and nsp8
SARS-CoV nsp7 and nsp8 are non-structural proteins that are involved in the replication and transcription of the SARS coronavirus. Nsp8 serves as an RNA primase, colocalizes with RdRP to replicate SARS-CoV, and is capable of de novo initiation. Nsp7 interactions with nsp8 boost RNA-dependent RNA polymerase (RdRp) and also increase RNA binding by nsp8.
Background
The outbreak of SARS, severe acute respiratory syndrome, in Hong Kong almost caused an pandemic and created a worldwide panic. From late 2002 to July 2003, there were about 8,273 cases and 775 deaths.[7] People who were infected reported muscle pain, headache, fever as well as respiratory symptoms such as cough and pneumonia.[8] The disease is known to infect older people, with mortality rate the highest for those over 50 years old.[8] SARS has not been eradicated and can return to infect the human population. Since no antibiotics, antivirals, steroids, and therapies are known to be effective against this viral disease, it has become a top priority for the government and public health scientists around the world to identify and develop vaccines and medicine to prevent another outbreak.
SARS coronavirus (SARS-CoV) was identified by a number of laboratories to be the main cause of SARS. In April 2003, Michael Smith Genome Sciences Centre, located in Vancouver, became the first lab in the world to sequence the coronavirus that caused SARS using samples from infected patients in Toronto.[4] The mapping of the SARS-CoV is a step forward to fighting SARS. With the sequence of SARS-CoV, scientists can begin to understand more about the coronavirus structure, function, and genome replication as well as begin antiviral research against SARS-CoV.
SARS-CoV is a positive strand RNA virus that causes respiratory and enteric disease.[3][6][Figure1] Viral replication of the coronavirus genome takes place in the cytoplasm. Infection by the coronavirus can alter cell cycle, transcription, translation pattern, and induce apoptosis as well as inflammation.[4]
Coronavirus (CoV) replication takes place in the cytoplasm and in a membrane-protected microenvironment.[2][9] The CoV genome has a 5’ methylated cap and a 3’polyadenylated tail. First, the S-protein of the CoV binds on the cell surface molecules.[2][9] It has been speculated that CoV either enters the host cell by fusion of viral and cell memebrane or by receptor mediated endocytosis. Once it enters the host cell, it directly produces its proteins and new genomes in the cytoplasm.[2][9] The CoV then synthesizes its RNA polymerase that recognizes and produces viral RNAs.[2][9] The RNA polymerase then synthesizes the minus strand using the positive strand as a template. The negative strand then serves as a template to transcribe smaller subgenomic positive RNAs, which are used to synthesize other proteins.[2][6][9] The negative strand also serves for the replication of new positive-stranded RNA genomes. The N protein of the CoV then binds the genomic RNA while protein M is integrated into the membrane of the endoplasmic reticulum.[2][9] The assembled nucleocapsids with helical twisted RNA that are formed after the binding of the N proteins enter the ER lumen and are encased with its membrane.[2][9] These viral CoV progeny are then transported by golgi vesicles to the cell membrane and enter the extracellular by exocytosis.[2][9]
RNA-dependent RNA polymerase
Genome replication and transcription in most RNA viruses are catalyzed by RdRp. RdRp, an enzyme, is an important protein that is encoded in the genomes of viruses containing RNA because it can catalyze the synthesis of the RNA strand complementary to a given RNA template[1][5]. Coronavirus replication and transcription are mediated by a protein complex with RdRP as a main component of the viral particles.[1][5] Furthermore, RdRP and other replicase proteins have been speculated to act as a starting replication machinery in the synthesis of CoV RNA by amplifying the genome before genome translation to improve the efficiency of virus infection.[1][5]
Nsp7 and nsp8
Nsp7 and nsp8 are two non-structural proteins that are found in SARS-CoV. Nonstructural proteins are encoded by a virus, but are not part of the viral particle. Nsp7 and nsp8 are important in the replication and transcription of SARS-CoV. The nsp(7+8) complex is a unique RNA polymerase that is capable of de novo initiation and primer extension.[4][10][12] Nsp8 is also known to colocalize with RdRP to copy the SARS-CoV genome.[10]
The discovery of the SARS-CoV has propelled extensive studies on its viral replication. Although the emergence of SARS-CoV has been extremely important towards finding a treatment for SARS, little is known about its replication machinery. However, scientists have learned more about SARS-CoV through the discoveries of nsp7 and nsp8, which have been identified to be involved in the CoV replication and transcription.[4][10][11][12] Furthermore, the interaction between nsp(7+8) complex is important for RdRp activities of the SARS-CoV genome.[10][12] By understanding the structures of these non-structural proteins, scientists are one step closer to decipher the mechanism of SARS-CoV genome replication and may even use them to develop new strategies for the prevention or treatment of SARS in animals and humans.
Structures of nsp7 and nsp8
Although there are other nonstructural proteins present in SARS-CoV, such as nsp9 and nsp10, the formation of the nsp(7+8) complex and its polymerase activity drive the RNA synthesis of SARS-CoV.[10][12] Without these proteins, SARS-CoV would be unable to replicate.
Nsp7 and nsp8 form the supercomplex of SARS-CoV. The transcription and replication machinery of SARS-CoV can be understood from this supercomplex. Yujia Zhai et al., was able to present the crystal structure of the hexadecameric nsp7 and nsp8 supercomplex of SARS-CoV at 2.4-A resolution.[12] This structure is considered to be the first to show interactions between CoV replication proteins as well as the architecture of the CoV replication and transcription machinery.[12]
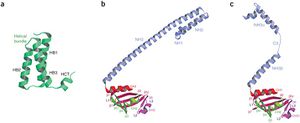
The structure of the supercomplex resembles a hollow cylinder with a central channel that has two handles protruding from opposite sides.[12] It is assembled from eight copies of nsp8 and is bound tightly together by eight copies of nsp7.[12] Nsp7 is an alpha helical protein of about 10kDA.[4][12] It has a single domain with a novel fold that consists of five helical secondary structure.[12] The interhelical side-chain interactions that stabilize and hold the helices together have residues that are predominantly hydrophobic.[12] Nsp7 localizes to cytoplasmic membrane structures, which are also known to be the sites of viral replication in infected cells.[4][12] SARS-CoV nsp7 dimerizes and interacts with other proteins such as nsp5, nsp8, nsp9 and nsp13.[4] Nsp7 central core consists of an N-terminal helical bundle (HB) that contains helices HB1, HB2, and HB3.[Figure 2] Its HB region is conserved and is known to interact with nsp8.[12] Nsp8 has a molecular mass of about 22kDa and is unique for coronaviruses.[4] The four monomers of nsp8 have two different conformations: nsp8I and nsp8II.[12] Nsp8I is described as a “golf-club” like structure that is composed of an N-terminal “shaft” domain that contains three helices (NH1-3) and a C-terminal “head” domain while nsp8II 2 has a similar head domain, but its shaft helix NH3 bends into two shorter helices.[12][Figure 2] The N-terminal is also revealed to be highly conserved, suggesting that its domain may have an important role in the interaction with other molecules or complexes.[12]
Structural interactions of nsp7 and nsp8
Interactions between nsp7 and nsp8
Interactions between nsp7 and nsp8 form the supercomplex.[Figure 3] Nsp8I and nsp8II bind tightly to nsp7, which cause the formations of two types of heterodimers, D1 and D2.[12] Hydrogen bonds are also formed between the side chains of nsp8 and nsp7.[12] There are two main sites in the supercomplex of the non-structural proteins. The first site is placed within the C-terminal complex of the shaft domain of nsp8.[12] Here, residues of the NH3/NH3 beta helix of the nsp8 and on the HB1 helix of the nsp7 form the hydrophobic core.[12] Site 2 is found in the CH1 helix of the nsp8.[12] The helices of nsp7, HB3 and HCT, form interactions with the CH1 helix.[12] The side chain and the main chain carbonyl group of nsp8 form hydrogen bonds with the main chain carbonyl group of nsp7.[12]
Interaction of the nsp7+nsp8 supercomplex with double stranded (ds)RNA
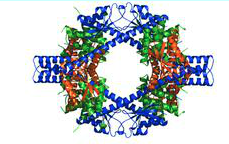
The structure and interaction of the nsp7-nsp8 supercomplex covey that its role is to bind nucleic acids. The inner channel of the complex has positive potential while the outer surface is covered by negative potential.[12] This charge distribution allows the phosphate backbone of nucleic acids to pass through the channel without electrostatic repulsion. The hollow cylindrical structure of the supercomplex also helps to facilitate CoV replication. Replication of the CoV occurs in the cytoplasm of infected cells and double-stranded (ds)RNA intermediate is required for the replication of SARS-CoV during RNA synthesis.[2][9][12] The hollow cylindrical of the supercomplex helps with the coronavirus replication, encircling, and stabilizing the dsRNA by holding the nascent and template strands together for transcription and replication.[4][10][12] Furthermore, the high conservation of nsp7 and nsp8 in coronaviruses indicates that they are general components for all coronavirus. The discovery that dsRNA and the supercomplex are natural binding partner can be useful for antiviral research for SARS. Peptides compounds can be designed to mimic the interactions between nsp7 and nsp8 to prevent the supercomplex from forming as well as to prevent the replication of SARS-CoV.[10][12] The structure of the supercomplex can also be used to design drugs to treat SARS.[10][12]
RdRp activities driven by nsp(7+8) complex
There are two RdRp activities that are believed to be involved in replication of SARS-CoV genome. The first activity involves the 106-kDa nsp12 while the second polymerase involves the 22-kDa nsp8. The first activity has the canonical viral RdRp motifs in its C-terminal part and employs a primer-dependent intiation mechanism.[10] The second polymerase activity, which involves nsp8, is unique for CoVs and is the only protein that is reported to be capable of de novo RNA synthesis.[10] De novo synthesis is the synthesis of complex molecules from simple molecules.

Figure 5.The nsp(7+8) complex has primer extension activity [10]
The supercomplex of nsp7 and nsp8 is known to be a unique RNA polymerase. Although nsp8 is capable of RdRp activities on its own, it is known to co-crystallize and form a unique hexadecameric ring-structure with nsp7. Aartijan J.W. te Velthuis et al., studied the relationship of nsp(7+8) complex to RdRp activities for the RNA virus SARS-CoV.[10] They found that exposure of nsp8’s natural N-terminal residue was important for its ability to associate with nsp7 as well also boosting its RdRp activity.[10]
nsp7 enhances activities of nsp8
To examine whether nsp7 improves nsp8’s function and capability to bind to RNA, measurement of dsRNA was measured using fixed concentration of nsp7 with either wild-type or mutant nsp8.[Figure 4] Without the presence of nsp8, no RNA binding was observed.[10] However, the presence of the formation of the nsp(7+8) complex created an increase in the number of bound dsRNA.[1][10] Furthermore, an increase in the bindng affinity for RNA was also observed for nsp8 RNA-binding mutants K58A and D52A.[10] These results showed that the addition of nsp7 must had evolved to enhance RNA binding by nsp8.
Primer extension activity was also found in the nsp(7+8) complex.[Figure 5] Further experiment showed the importance of nsp7 to nsp8 and its primer extension activity. The nsp7 was shown to directly increase the primer extension activity that was driven by the nsp8.[1][10] The comparison of the two enzyme complexes found that the primer extension activity of nsp8 alone was less than 2-fold lower than when both nsp7 and nsp8 were present at equal molarity.[1][10]
Conclusion
Recent studies on the structures of nsp7 and nsp8 have provided important insight on SARS-CoV. Scientists have discovered that nsp7 and nsp8 do not only interact with each other, but that nsp8 is dependent upon nsp7. These interactions and dependency are important for the replication of the SARS-CoV genome. Furthermore, the hollow cylindrical structure of the supercomplex of nsp7 and nsp8 is important for the facilitation of the CoV replication.[4][10][12] Without this structure, the RdRp activities are limited.[1][4][10][12]
nsp7 enhances activities of nsp8
Although the discovery of these non-structural proteins has allowed scientists to examine the replication mechanism of SARS-CoV, more experiments are needed in order to better understand the functions of nsp7 and nsp8. For example, there are two RdRp activities, with either nsp 12 or nsp(7+8), that are involved in the replication of SARS-CoV genome.[10] However, not much is know whether these two polymerase activities are within the same enzyme complex or whether they can influence each other's activity.
Since nsp7 and nsp8 are known to be important to the replication of SARS-CoV, it is also important for future research to study how they can be used to treat diseases that are caused by this virus. Furthermore, it is crucial to begin intensive antiviral research using nsp7 and nsp8 as models to develop drugs since SARS has not been eradicated and a potential outbreak may occur in the future.
References
1. Butcher, S.J., Grimes, J.M., Makeyev, E.V., Bamford, D.H., and Stuart, D.I. (2001). A mechanism for initiating RNA-dependent RNA polymerization. Nature 410, 235–240.
2.Enjuanes, L., Almazán, F., Sola, I., and Zuñiga, S. (2006). Biochemical Aspects of Coronavirus Replication and Virus-Host Interaction. Annual Review of Microbiology 60, 211–230.
3. Hunt, Richard. "CORONA VIRUSES, COLDS AND SARS." Microbiology and Immunology. University of South Carolina School of Medicine , 076 Apr 2010. Web. 25 Mar 2013.
4. Lal, Sunil K. Molecular Biology of the SARS-Coronavirus. New Delhi: Springer , 2010. eBook.
5.Nogales, A., Marquez-Jurado, S., Galan, C., Enjuanes, L., and Almazan, F. (2012b). Transmissible Gastroenteritis Coronavirus RNA-Dependent RNA Polymerase and Nonstructural Proteins 2, 3, and 8 Are Incorporated into Viral Particles. J Virol 86, 1261–1266.
6.Sawicki, S.G., and Sawicki, D.L. (1998). A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440, 215–219.
7.Surjit, M., Liu, B., Kumar, P., Chow, V.T.K., and Lal, S.K. (2004). The nucleocapsid protein of the SARS coronavirus is capable of self-association through a C-terminal 209 amino acid interaction domain. Biochem. Biophys. Res. Commun. 317, 1030–1036.
8.Trivedi, Manish N. "Severe Acute Respiratory Syndrome (SARS)." Medscape Reference. University of South Carolina School of Medicine . Web. 25 Mar 2013.
9.Van Hemert, M.J., Van den Worm, S.H.E., Knoops, K., Mommaas, A.M., Gorbalenya, A.E., and Snijder, E.J. (2008a). SARS-Coronavirus Replication/Transcription Complexes Are Membrane-Protected and Need a Host Factor for Activity In Vitro. PLoS Pathog 4, e1000054.
10.4. Velthuis, A.J.W. te, Worm, S.H.E. van den, and Snijder, E.J. (2011). The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucl. Acids Res.
11. Xiao, Y., Ma, Q., Restle, T., Shang, W., Svergun, D.I., Ponnusamy, R., Sczakiel, G., and Hilgenfeld, R. (2012). Non-structural Proteins 7 and 8 of Feline Coronavirus Form a 2:1 Heterotrimer that Exhibits Primer-independent RNA Polymerase Activity. J. Virol.
12. Zhai, Y., Sun, F., Li, X., Pang, H., Xu, X., Bartlam, M., and Rao, Z. (2005). Insights into SARS-CoV transcription and replication from the structure of the nsp7–nsp8 hexadecamer. Nat Struct Mol Biol 12, 980–986.
Edited by (Amy Tran), a student of Nora Sullivan in BIOL187S (Microbial Life) in The Keck Science Department of the Claremont Colleges Spring 2013.