Saprospira grandis
A Microbial Biorealm page on the genus Saprospira grandis
Classification
Higher order taxa
Domain: Bacterial
Phylum: Bacteroidetes
Class: Sphingobacteria
Order: Sphingobacteriales
Family: Saprospiraceae
[5]
Species
NCBI tax ID: 694433
Genus: Saprospira
Species: grandis
[5]
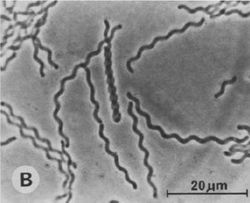
Description and significance
Saprospira grandis is a gram-negative, marine, multicellular, filamentous flexibacterium. It is motile; but it does not swim, it glides, which is why it has been classified as a flexibacterium. The cells that have been observed appear peach colored and contain a unique carotenoid, saproxanthin, which is thought to be photoprotective against damage by solar ultraviolet light. Another striking feature is the abundant presence of rod-shaped structures within the cells known as “rhapidosomes.” Currently, no one knows the physiological purpose of these structures or how they are synthesized and regulated. Saprospira grandis are free-living, commonly found in marine littoral sand and coastal zones in various locations around the world. They prey on other bacteria by trapping and devouring them. It is significant because it is known for devouring bacteria and also has been shown to digest algae by the same process. This makes this bacterium important because it is useful in preventing harmful algal blooms. [1,3]
Genome structure
While bacteria of the genus Saprospira are studied quite extensively, genome information is lacking thus far. Therefore, it is of interest to obtain the complete genome sequence of S. grandis to determine its metabolic potential, predatory lifestyle, and genes that encode proteins involved in rhapidosome formation. S. grandis str. Lewin, was the first member of the Saprospiraceae family to have its complete genome sequenced. The complete genome sequence comprises a chromosome of 4.35 Mbp and a plasmid of 54.9 Kbp. The organism has incomplete pathways for the biosynthesis of nine essential amino acids but encodes a large number of peptidases. The S. grandis genome encodes ten sensor globin-coupled rsbR genes thought to be essential for stress response and the presence of such sensor globins in Bacteroidetes is unprecedented. Genome analysis revealed a total of 429 spacer sequences within the three clustered regularly interspaced short palindromic repeat regions in the genome and this number is the largest among all the Bacteroidetes species sequenced to date. [5]
Cell and colony structure
The filaments on Saprospira grandis are about 1 micrometer wide and indefinite in length (5-500 µm), with a shallow helical pitch of 5-10 micrometers. Which consist of cells 1-5 micrometers long and separate as they die. They have solid substrata, which is used for gliding, at 2-5 micrometers sec -1. The cells are peach colored and contain carotenoid saproxanthin, which may be photoprotective against damage by solar UV light. They have an abundant presence of rod-shaped structures within the cells known as “rhapidosomes.” It has already been postulated that rhapidosomes are defective phage tails that could be bacteriocins similar to the pyocin R bacteriocin of Pseudomonas aeruginosa. If this is true, the rhapidosomes should play a critical role in the ability of Saprospira to consume other bacteria. [1,3]
Metabolism
S. grandis is a chemoorganotroph, which means they oxidize the chemical bonds in organic compounds as their energy source. Chemoorganotrophs also attain the carbon molecules that they need for cellular function from these organic compounds. They are capable of decomposing many different biomacromolecules. The organic compounds that they oxidize include sugars, fats, peptides and proteins. S. grandis are strictly aerobic. [6]
Ecology
Saprospira grandis is a marine bacterium. They are mesophilic with their optimum temperature being between 25-30 degrees Celsius, and require a neutral pH. They can grow well at 30ºC but can survive at 40ºC for several hours. Saprospira grandis are free-living bacterium found in marine littoral sand and coastal zones in various locations around the world. They contribute to the environment by consuming algae in helping to prevent harmful algal blooms. [1,3]
Pathology
S. grandis prey on other bacteria and protists. Unsuspecting bacteria swimming nearby become trapped when the tips of their flagella become stuck in the mucilage on the saprospira filament. No chemotaxis seems to be involved, just approaching too closely. Dozens of live bacteria still spinning on their axes become attached to a single filament, eventually to cover its surface. Extracellular enzymes secreted by the saprospira finish them off. Not all motile bacteria qualify as prey. For example, non-marine but flagellated cells such as E. coli aren't prey, and only 7 out of 25 marine members of the Vibrionaceae could be caught and destroyed. [2,4]
References
[1] A.S. Delk, C.A. Dekker. "Characterization of Rhapidosomes of Saprospira Grandis." Volume 64, Issue 1. Pages 287-288. Berkeley, California: Department of Biochemistry, 2012. Print.
[2] Elio. "Small Things Considered." : Of Fly Paper, Esperanto,Algae, Poems, and Polar Bears: In Memory of Ralph Lewin. 08 Jan. 2009. Web. 08 Mar. 2012. <http://schaechter.asmblog.org/schaechter/2009/01/of-fly-paper- esperanto-algae-poems-and-polar-bears-in-memory-of-ralph-lewin.html>.
[3] Lewin, R.A. Microbial Ecology. "Sapropsira Grandis: A flexibacterium." Volume 34 Pages 232-236. Springer-Verlag New York Inc. February 6, 2012.
[4] Sangkhobol, Vullapa, and Victor B. D. Skerman. Current Microbiologyi. "Saprospira Species-Natural Predators." Volume 5, number 3. Pages 169-174. SpringerLink. Springer Science Business Media. Web. 07 Mar. 2012. <http://www.springerlink.com/content/n570722rk1728h6j/>.
[5] Saw, Jimmy H.W. "Standards in Genomic Sciences." Www.standardsingenomics.org. 05 Mar. 2012. Web. 07 Mar. 2012. <http://standardsingenomics.org/index.php/sigen/view/sigs.2445005/684>.
[6] Williams and Wilkins, William. "Bergey’s Manual of Determinative Bacterialology." 9th ed. Lippincott, 1994. Print.
Edited by Renee Nicholas of Dr. Lisa R. Moore, University of Southern Maine, Department of Biological Sciences, http://www.usm.maine.edu/bio
