Sphingomonas wittichii RW1
Classification
Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae
Species
Sphingomonas wittichii RW1
Description and Significance
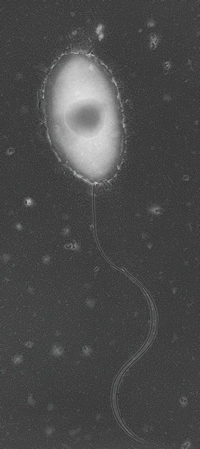
The S. wittichii bacterium is Gram negative, rod-shaped, monotrichous, and asporogenous. It was isolated from the Elbe River in Germany, and is noted for its ability to degrade dioxins, chemicals that are produced as byproducts in industrial processes, particularly processes that involve combustion. Other Sphingomonas bacteria have been found in soil, and indeed the RW1 strain that contains the dioxin-degradation pathway can be grown on agar plates, even though it was isolated in an aquatic environment.
Along with the ability to mineralize the dioxin backbone, S. wittichii is also capable of degrading other xenobiotics, such as toluene, fluorobenzoate, styrene, and carbazole [8]. These degradation pathways make S. wittichii a key organism in the bioremediation of dioxin and other pollutants.
Genome Structure
| Name | Topology | Length (Mbp) | GC content | % Coding | Proteins |
|---|---|---|---|---|---|
| Chromosome | circular | 5.4 | 68.4% | 91% | 4850 |
| Plasmid pSWIT01 | circular | 0.31 | 64.1% | 85% | 285 |
| Plasmid pSWIT02 | circular | 0.22 | 61.2% | 83% | 210 |
Cell Structure, Metabolism and Life Cycle
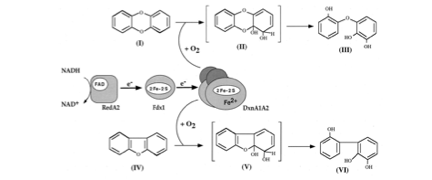
Cell Structure – S. wittichii RW1 is, as mentioned above, a rod shaped bacteria with one flagellum. The cell is considered motile. RW1 is the only known organism to carry the dioxin dioxygenase enzyme [1], which is an enzyme that initiates the oxidation of the dioxin molecule by adding two oxygen atoms. This is probably the most sought after characteristic of the organism [2]. The dioxin dioxygenase is made of three subunits encoded by the dxnA1, dxnA2 and ferridoxin genes. Interestingly, the genes that encode the electron supply system (a flavoprotein reductase, redA2, and ferredoxin, fdx1) for the enzymes are located on one of the other two genomic sections [2]. Some have proposed that this non-grouping of the required genes indicates that the bacteria either recruited the genetic code from other organisms, or the dioxin degrading machinery is in the process of evolution [4]. Dioxin dioxygenase attacks the dioxin molecule at the oxygen between the two aromatic rings (angular attack), whereas most other dioxygenases attack at carbons other than the ones next to the central oxygen. Dioxin dioxygenase is also unique in that its electron supply system uses a putidaredoxin-type ferridoxon instead of the common Rieske-type ferridoxin [2].
Figure 1: Dioxin degradation pathway, from Armengaud, 1998 [2].
Metabolism – RW1 is an aerobic organoheterotroph. The organism uses organic carbon as its source of energy and carbon and O2 is its terminal electron acceptor. What is fascinating about this organism is that it can use a diverse array of aromatic organic compounds as its energy and carbon source, including some extremely recalcitrant (degradation resistant) compounds, including dibenzo-p-dioxin (DD, or dioxin), some chlorinated and polychlorinated DD, dibenzofuran (DF), and many other compounds [5]. S. wittichii RW1 has been the subject of many studies trying to show all the compounds the bacteria can degrade, including many different congeners (species) of chlorinated DD. A recent study has shown that the bacteria can degrade one congener of dioxin that has 6 chlorine substitutions on the compound (1,2,3,4,7,8-hexachlorodibenzo-p-dioxin). The bacteria can degrade multiple congeners of dioxin with one or two chlorine substitutions [6].
Life Cycle – The generation time of RW1 is 8 hours and 5 hours when the organism is grown with DD and DF respectfully [5]. RW1 produces a type of sphingoglycolipid (SGL) called glucuronosyl ceramide (SGL-1). This is a relatively unique lipid, and its discovery is what led to the creation of the genus ‘Sphingomonas’. S. wittichii RW1 is one of just a few Sphingomonas species to produce the novel sphingoglycolipid, galacturonosyl ceramide (SGL-1') in addition to the more common SGL-1 [7].
Ecology and Pathogenesis
Symbiosis and Pathogenesis – Currently, there is no research being done on symbiotic relationships between S. wittichii and other organisms, nor has S. wittichii been found to be pathogenic.
Contributions to environment – As mentioned before, S. wittichii is an important organism in bioremediation and in understanding the degradation process of various xenobiotics. One of the systems that S. wittichii has been tested in for removal of dioxins is in fly ash—a residue generated in the combustion of coal. When fly ash was collected from municipal waste incinerators and exposed to S. wittichii, three-quarters of the toxic polychlorinated dibenzo-p-dioxins (PCDDs) were removed [9]. This is a significant step forward, of course, since dioxins are so susceptible to bioaccumulation [7].
References
1. Halden RU, Colquhoun DR, and ES Wisniewski, 2005. Identification and phenotypic characterization of Sphingomonas wittichii strain RW1 by peptide mass fingerprinting using matrix-assisted lazer desorption ionization-time of flight mass spectrometry. Appl Env Microbiol, 71:2442-2451.
2. Armengaud J, Happe B, and KN Timmis. 1998. Genetic analysis of dioxin dioxygenase of Sphingomonas sp. strain RW1: catabolic genes dispersed on the genome. J. Bacteriol. 180:3954–3966.
3. Bunz PV and AM Cook. 1993. Dibenzofuran 4,4a-dioxygenase from Sphingomonas sp. strain RW1: angular dioxygenenzyme system. J. Bacteriol. 175:6467–6475.
4. Nojiri H, Habe H, and T Omori. 2001. Bacterial degradation of aromatic compounds via angular dioxygenation. J. Gen. Appl. Microbiol. 47:279-305.
5. Wittich RM, Wilkes H, Sinnwell V, Francke W, and P Fortnagel. 1992. Metabolism of dibenzo-p-dioxin by Sphingomonas sp. strain RW1. Appl. Environ. Microbiol. 58:1005-1010.
6. Nam IH, Kim YM, Schmidt S, and YS Chang, 2006. Biotransformation of 1,2,3-tri- and 1,2,3,4,7,8-hexachlorodibenzo-p-dioxin by Sphingomonas wittichii Strain RW1 Appl. Environ. Microbiol. 72:112-116.
7. US DOE Joint Genome Institute. "Sphingomonas wittichii RW1" [online] http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=detailssearch&term=txid392499%5Borgn%5D%20AND%20pt_default%5Bprop%5D%20.
8. “Sphingomonas wittichii” KEGG Genome. http://www.genome.jp/kegg-bin/show_organism?menu_type=pathway_maps&org=swi.
9. In-Hyun Nam, et. al. 2005. Biological removal of polychlorinated dibenzo-p-dioxins from incinerator fly ash by Sphingomonas wittichii RW1. Water Res. 39:4651-4660.
10. Entrez Genome Project. “Sphingomonas wittichii” [online] http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=17343.
Author
Page authored by Andrea Johnson and Timothy Johnson, students of Prof. Jay Lennon at Michigan State University.