Staphylococcus aureus: USA:300
A Microbial Biorealm page on the genus Staphylococcus aureus: USA:300
Classification
Domain: Bacteria
Phylum: Firmicutes
Class: Bacilli
Order: Bacillales
Family: Staphylococcaceae
Genus: Staphylococcus
Species: S. aureus
(Strain: USA:300)
Description and significance
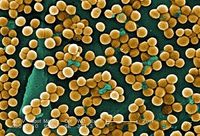
Staphylococcus aureus: USA:300 is a strain of gram-positive coccus bacteria responsible for Methicillin-resistant Staphylococcus aureus (MRSA), or Staph infection in humans. This strain of S. aureus is resistant to β-lactam antibiotics. When cultured, this bacteria appears as golden clusters. The golden color is the result of a carotenoid pigment that protects the bacteria against host-immune system reactive oxygen species, and adds to the bacteria's virulence. S. aureus is a facultative anerobe, which can be found in a wide variety of locations such as soil, human skin, and public places like hospitals and prisons. This strain of S. aureus: USA 300 was first identified in 1998, and is thought to be the primary causal strain of community-acquired Staph infections throughout the United States, Canada and Europe. In 2006 the CDC reported that 64% of MRSA isolated from infected patients were of the USA 300 strain. This bacteria contains the cytotoxin Panton-Valentine leukocidin (PVL), which targets leukocytes. It also contains modulin, which is a phenol-soluble peptide (PSM) that is capable of lysing neutrophilic granulocytes. These toxins cause rapidly-progressing fatal conditions such as necrotizing pneumonia and faciitis and severe sepsis. USA:300 causes an estimated 20-40 thousands deaths annually and worldwide.
Genome structure
The genome of S. aureus: USA:300 was first sequenced in 2006. It contains a single, circular chromosome. This strain was found to have a genome of 2,872,769 base-pairs (bp) and 2,560 genes. It also contained a 3.1 kilobase (kb) plasmid and 27kb plasmid. As expected the genome contained genes encoding for the cytotoxins PSM and PVL, as well as an Arginine catabolic mobile element or (ACME) gene which is thought to have been acquired by USA:300 from S.epidermidis by horizontal gene transfer. The presence of ACME helps to explain the prevalence of Staph infections of the skin.
Cell structure, metabolism & life cycle
S. aureus: USA:300 is a facultative anaerobe, meaning that it is capable of aerobic respiration in the presence of oxygen, and in an anoxygenic environment can switch to fermentation. It utilizes the pentose cycle in carbohydrate breakdown, and glycolysis in glucose metabolism. It is not known to use the Entner-Doudoroff pathway. Interestingly, in iron-deficient environments, S. aureus has been found to redirect its central metabolsim to sequester nutrients, helping it to survive in hostile host environments.
S. aureus: USA:300 is a gram-positive bacteria, and contains a thick layer of peptidoglycan. The cell walls also includes murein, teichoic acids, and surface proteins. The murein layer is extremely complex and is composed of glycan and oligopeptide chains arranged in a plane perpendicular to the plasma membrane. The oligopeptide chains are thought to confer a "zig-zag" arrangement and the glycan strands are considered to be "zippering". Together, these form a macromolecular sacculus surrounding the entire cell. The surface proteins play a multitude of different roles. They contain a class of surface protein adhesions, known as MSCRAMMs which assist the bacteria in adhering to host-immune cells.
Ecology (including pathogenesis)
As previously mentioned S. aureus: USA:300 is commonly found in soil, public places and on epidermis of humans and other animals. A surprising 30% of humans are thought to contain the bacteria on their skin, with only a fraction of that population ever being infected by the pathogen. It is commonly found in high person-to-person contact areas such as prisons, hospitals, schools and sports-fields. There has also been frequented reports of S. aureus: USA 300 outbreaks in homosexual men in the United States and Europe.
The organism is pathogenic for several reasons. It posses two main toxins, which destroy host-cell immune response. The first of these toxins is Panton-Valentine leukocidin (PVL). It works as a β-pore-forming toxin that creates unregulated pores within the membrane of host-cells. The result of this infection leads to lesions of the skin or mucosa. The second main virulence factor are the phenol-soluble modulins (PSMs). These are a group of peptides which are roughly 20 amino acids in length. They recruits and lyses neutrophils, and may be one of the leading causes of sepsis to the host system.
Although the bacterial strain is common, S. aureus: USA: 300 typically infects those who are immunocompromised or in a susceptible environment (as before mentioned). Infections will often enter the body through a small cut or sore. Other symptoms associated with USA:300 are pneumonia, necrotizing fasciitis, endocarditis, and bone and joint infection. Treatment is difficult since this strain of bacteria is resistant to most β-lactam antibiotics, these include; penicillin, methicillin, dicloxacillin, nafcillin and oxacillin. β-lactam antibiotics work by binding to penecillin-binding-proteins (PBPs), which inhibit cell-wall synthesis of peptidoglycan. β-lactam resistant antibiotics alter the binding of the antibiotic to PBPs.
Interesting feature
One of the distinguishing features of S. aureus: USA:300 are the components of its 27 kbp plasmid (pUSA300HOUMR) and its 2.8 million bp chromosome. The genome contains SCCmec, which confers methicillin resistance, while the plamid encodes resistance to a whole range of antibiotics including; β-lactams, marcolides, aminoglycosides, streptothricin and bacitracin. All of the pathogenic genes or "islands" are found in the chromosome, these include genes for; enterotoxins, exotoxins, leukotoxins, hemolysin, exfolatin and epidermin. There is a great deal of sequence homology in pUSA300HOUMR and the plasmid pSERP from Staphylococcus epidermidis, a non-pathogenic species of Staphylococus which inhabits normal skin flora. Therefore it is thought that S. aureus acquired its plasmid from S. epidermidis, and that the latter gained antibiotic resistance because it inhabits skin and is frequently in contact with antibiotics secreted by sweat.