Streptococcus mutans: More than Oral Pathogens
By: Jordan Wehner
Introduction
Image of Gran stained
History
Our genetic code isn’t the only thing that is similar to chimpanzees; we also have oral microbes in common. Specifically, we share the firmicute Streptococcus mutans (Fig. 1). These pathogenic microbes are part of the bacteria kingdom and were first isolated from human cavities (caries) in 1924 by Clarke [1]. They received their name (S. mutans) because Clarke believed that the microbes were a mutant version of Streptococcus. These microbes are found in all Homo sapiens and are the main cause of dental decay. As much as 90 % of the world is affected by this pathogenic microbe [2].
Serotypes
Like many species of bacteria, S. mutans have various strains and serotypes. Serotypes are microorganisms that share a specific substance which causes an immune response. Four major serotypes have been discovered thus far. The serotypes are labeled as c, e, f, and k.Sserotype c is considered to be the most common serotype found in plaque, making up about 80 % of the microbes population. [3] The other three serotypes (e, f, and k) are less common. Up to 20 % of the S. mutans are considered to be e. The smallest groups, f and k, are thought to make up 5 % of the microbe population. Interestingly, serotype k has only been isolated from Japanese individuals. Recent research has found that e, f, and k may play a hazardous role in infective endocarditis (e and f) and hemorrhagic strokes (k) even though they only make up only about a quarter of the population.
Genome Structure
Currently, full genome sequences of only two serotypes of S. mutans are known. The first to be sequenced was the c serotype. In particular, two c strains have been fully sequenced, UA159 and NN2025. [4],[5] More recently, the genome of a serotype k was sequenced in Japan in order to learn more about its relationship with hemorrhagic stroke. [6]
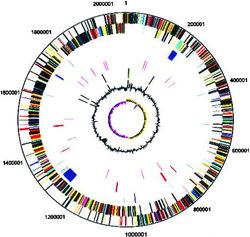
The sequencing of the first S. mutans was published in 2002 and was the UA159 strain (c serotype). [4] It was isolated and sequenced in the United States. It was composed of a single circular genome with 2,030,0936 base pairs (Fig. 2). When sequencing the DNA they found that 36.82 % of the content was GC. This low GC content is consistent with the environment that S. mutans live in. The mouth, on average, is 20 – 40 oC and with a pH of ~ 7. These conditions are important for preserving the bacterial DNA; if the oral cavity was more extreme a higher GC content would be needed to keep from destroying the DNA. The genome consists of 1,963 open reading frames (ORFs) organized so that both DNA replication and transcription are in the same direction. About a 1/5 of the ORFS are unique to the UA159 genome. Five rRNA operons and 65 tRNA sequences are located in the genome.
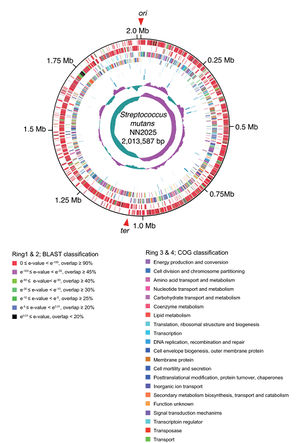
The strain NN2025 was the next strain to be fully sequenced and was published in 2009.[5] This strain was isolated in Japan and was found to contain 2,013,597 base pairs (Fig. 3). The GC content of this strain was 36.85 %, very similar to that of the UA159 strain. The NN2025 strain contained 1,895 ORFs, 68 less than the UA159 strain. While the genome may be slightly smaller, the genome also consists of five rRNA operons and 65 tRNA sequences. Both genomes contain sequences for the production of proteins and enzymes involved in the fermentation of sugars. When the sequences were compared their appeared to rearrangement in the genome.
With only about five percent of the S. mutans serotypes being k, it is not surprising scientists have not isolated and sequenced a k serotype until 2012. [7] The strain was LJ32 and was isolated in Japan. This new serotype genome, like both the c serotypes, is composed of one circular genome. This genome consists of between 2,015,626 base pairs, similar to that of the c serotypes. The GC content in this strain is slightly higher than that of the c serotypes, 37.05 %. This strain contained the same rRNA operons and tRNA sequences. When the LJ23 sequence was paralleled with the UA159 and the NN2025, there was genomic rearrangement similar to that of the NN2025. This is believed to be due to the place of isolation, since the UA159 strain was isolated in the United States and NN2025 and LJ23 were both isolated in Japan.
Metabolism
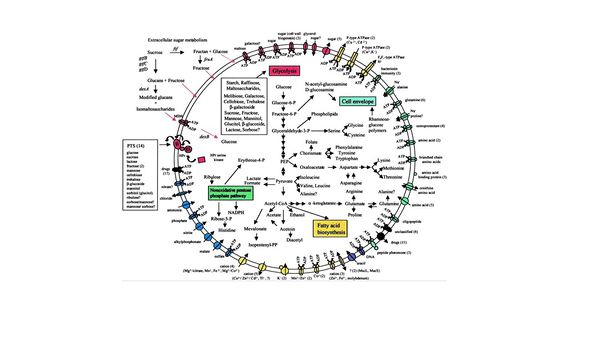
Through the sequencing of the UA159 strain, scientists have been able to further understand the metabolic pathway involved in sugar fermentation without oxygen (Fig. 4).[4] Scientists believe that S. mutans The major sugar consumed by S. mutans is sucrose. The sugars metabolized by the microbe are converted into polysaccharides composed of D-glucose monomers by glucosyl transferase [8]. These glucans allow the microbes in a colony to stick to each other along with the surface of the tooth. Many dentists refer to this as plaque. The glucans are then modified and cross the membrane by an ATP-driven transporter and broken back down to glucose. From here the sugars undergo glycolysis forming an end product pyruvate. The fermentation of pyruvate is S. mutans main source of energy. The pyruvate is fermented and the products are ethanol, formate, acetate, and lactic acid. Along with fermentation, the cell has many other uses for intermediates and pyruvate, such as fatty acid synthesis, cell upkeep, and a non-oxidative pentose phosphate pathway. The acid products of the fermentation are then deposited in the environment surrounding the cell within a colony. By doing this, S. mutans allow themselves to keep their niche in the oral cavity by preventing other microbes from growing in the environment. S. mutans are able to keep homeostasis by having membrane bound proteins that uphold a nearly neutral intracellular pH.
Acid Production and Teeth
The fermentation and production of acid not only inhibits competition but also causes dental caries [8],[9]. With hundreds of microbes living in every individual’s mouth it is hard to believe that one microbe is the major contributor to dental caries throughout the world. As the world continues to produce and consume more and more refined sugars, dental hygiene becomes more important. As we continue to consume more sugars we begin to fuel the production of acids and accelerate colony growth, which leads to the formation of dental plaque. The fermentation of the sugars to acids drops the environmental pH. It does not lower the pH of the entire mouth but just the area in which the colony inhabits. This is very important in order to understand how these microbes affect dental health. In order for the enamel and dentin of the tooth to remain rigid, the tooth must be in an environment above a pH 5.5. When the colony ferments, the pH drops below 5.5, and the acids secreted by the microbes begin to demineralize the enamel, causing the tooth to “rot.” The enamel is protects the dentin, root canal, and pulp chamber. The root canal allows for blood vessels and nerves to enter and exit the pulp chamber, keeping the tooth healthy. When the S. mutans demineralize the enamel, it begins to also irreversible destroy the dentin and could infect the pulp chamber. From there the microbes could enter the blood stream and travel throughout the body and kill the tooth. Once S. mutans are in the blood stream, scientists have discovered that the microbes may be more harmful than expected.
S. mutans "Attachment" to Your Heart
Recently, researchers have been trying to find a reason for the correlation between dental health and cardiovascular health [10]. By observing what microbes are found at cardiovascular disease sites, scientists have found that S. mutans are often prevalent at the location where the cardiovascular disease took place. The microbes are able to access these areas by entering the body during dental procedures (root canals and tooth extractions). Another means of access is through a rotting or infected tooth. If enough S. mutans enter the blood stream, it is likely that the microbes could reach the heart tissue. From here the microbes may attach to the endothelial cells using a special collagen binding protein. By binding to endothelial cells, they could infect the surface of the heart or create blockages in the heart's arteries.
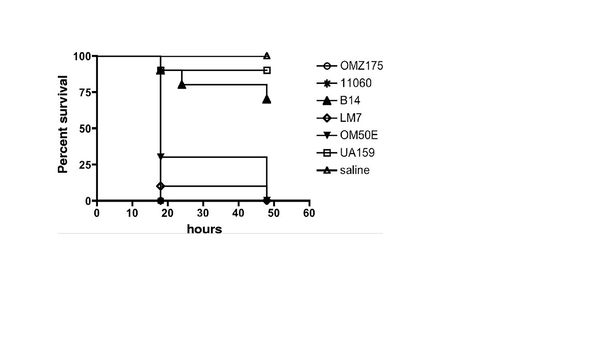
Researchers believe that the main contributor to this issue is S. mutans. These microbes can contain a collagen/laminin-binding protein (cnm) located on the surface of the microbe. Luckily, the serotypes that contain these proteins are the rarer serotypes e, f, and k. This research group explored binding activity and invasive character of two serotypes, e and f. The strains used were OM50E (e), LM7 (e), B14 (e), OMZ175 (f), and NCTC11060 (f). OM50E, LM7, B14, and OMZ175 were isolated form dental plaque form American patients. NCTC11060 was the only strain isolated form the blood of a patient. The k serotype was not studied because of their isolated discovery in Japan and because they are considered the rarest serotype.
To test this theory, strains were isolated from patients and cultured using a brain heart infusion medium. Researchers were able to prove the serotype of the strains using PCR techniques. Once the serotypes were known, they were tested for adherence and invasion of human coronary artery endothelial cells, ability to infect Galleria mellonella (wax worms), ability to form biofilms, and their ability to survive in blood. Previous work has shown that Galleria mellonella are ideal models when studying infectious diseases effecting mammals.
The strains specified above were chosen from a group for further studying because they were the five strains that showed invasive properties, contained the cnm gene, and expressed the protein (Fig. 5). There appeared to be no c serotype strains that had the cnm protein or invasive property. Some of the e serotypes failed to contain the cnm gene. The f serotypes appeared to be most likely to be invasive after incubating in human coronary artery endothelial cells. To test the invasive properties the strains were introduced to Galleria mellonella and saline was used as a control. UA159 was used for a baseline and to test if the cnm negative S. mutans may also have a similar effect as the cnm positive strains. As expect all the cnm positive strains showed an increased mortality rate within the Galleria mellonella colonies. The c serotype was similar to that of the control.
In order to test the original hypothesis that the cnm protein is a contributor to the strains ability to attach and invade human coronary artery endothelial cells, mutant genes were cultured. The cnm gene was knockout in the OMZ175 strain creating OMZ175-cnm. They first tested the binding ability of the cnm knockout. The UA159 strain was used as control. The results showed that the cnm knockout strain's ability to bind to the human coronary endothelial cells decreased drastically. The attachment was similar to that of the c serotype (UA159). The cnm knockout strain was then administered to Galleria mellonella to test if they would have any effect on mortality rates. The results of the OMZ175-cnm strain were similar to the c serotype (UA159). The other four strains an identical test was done and the results were all similar. By knocking out the cnm gene, the binding and invasive properties were similar to the c serotype and appeared to have no effect on the Galleria mellonella.
The researchers then tested how knocking out the cnm gene affected S. mutans ability to form plaque and survive. the cnm knockout strains did not appeared to have an altered ability to form biofilms. It did not appear that the cnm gene affects the ability to survive and cultivate within blood. Through these results the group was able to provide enough evidence to support their hypothesis that the cnm protein plays an important role in the binding and invading of human coronary artery endothelial cells. They researchers also believe that the cnm protein may be an efficient way to prevent S. mutans related cardiovascular infections. This could be done by fluorescently marking the cnm cells and screening for the pathogenic microbes during check-ups.
The Correlation Between S. mutans and Hemorrhagic Stroke
As scientists go on studying S. mutans, they continue to discover how pathogenic this microbe might be. The same year those researchers discovered a connection between S. mutans and cardiovascular disease, they also unearthed a link between these pathogenic microbes and hemorrhagic stroke [6]. The idea that the S. mutans could cause a stroke would not be far fetched. These microbes have access to the blood stream, through methods already depicted in the above section. Once they have access to the blood stream, the pathogenic microbe could find its way to the brain. In fact, doctors have found S. mutans, serotype k, in some Japanese patients treated for the narrowing of blood vessels in the brain. With this knowledge it is not difficult to believe that k serotypes may play a role in hemorrhagic strokes.
This group tested the effects of the S. mutans on the hemorrhages within mice. They used the following strains: MT8148 (c), TW871 (k), TW295 (k), SA53 (k), and LJ32 (k). The individual strains were injected into mice via a blood vessel in the tail. A control of saline was used and the MT8148 strain was used to test if the most common serotype (c) had similar effects. Shortly after the injection a blood vessel in the mouse’s left cerebral hemisphere was damaged using photo-irradiation. Three hours after the blood vessel was damaged, a sample was taken and the microbes were detected in the area of the damage. Then twenty-four hours later, the area of the hemorrhage was measured for each strain. Interestingly, the strains were only found in one area of the body. The results showed that the k serotypes were only found in the area of the hemorrhage, in the left hemisphere of the brain. The hemorrhage area was also determined to be dramatically larger for the k serotypes. The c serotype had an identical hemorrhage area as the control. In order to determine if damage was necessary for hemorrhaging, the k serotypes were injected to mice without damaged cerebral blood vessels. They were then monitored and no hemorrhaging was observed. Thus, the idea that the S. mutans could cause hemorrhaging without the damage of a blood vessel is unlikely.
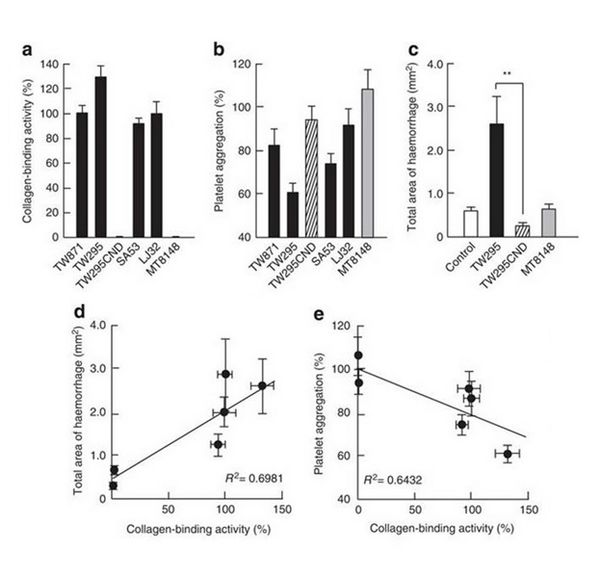
The researchers were then determined to identify the reason behind the increased cerebral bleeding. They noticed through testing, that the k serotypes were able to induce a protein known as MMP-9. This is important because MMP-9 has shown to increase the amount of bleeding in previous studies [11]. Also, in older researcher papers it has been discovered that certain strains and certain serotypes contain the collagen protein cnm discussed above. All four k serotype strains contained the cnm gene and expressed it. This could greatly influence the ability for platelet activation and the clotting preventing further bleeding. To understand this, it is important to understand how clotting works. For platelet activation and clotting in a damaged blood vessel to occur, the platelets must bind the collagen fibers that are only present after the vessel is damaged. The hypothesis was that these cnm proteins were binding to the collagen fibers present at the site of damage. Thus, the collagen binding, by the S. mutans inhibits the interaction between the collagen and the platelets. Without the platlet collagen interaction it allowed for the enhanced hemorrhaging. To test this hypothesis, a cnm knockout mutant strain of TW295 was created (TW295CBP). The four k serotype strains, the TW295CBP strain, and the c serotype were tested for their ability to bind collagen fibers. All four cnm positive strains showed close to 100% binding activity. The TW295CBP strain and the c serotype showed little to no binding activity. Then the strains ability to inhibit platelet activation and clotting ability was tested. The four cnm strains showed a reduction in platelet activation and clotting (10 – 40 % reduction). The TW295CBP strain and c serotype showed no significant difference in activation or clotting. TW295CBP was then tested for its effects on hemorrhage area. The results were similar to that of the control and c serotype. The TW295 strain produced five times the hemorrhage area than knockout strain. Figures 6 show the medium strength correlation of S. mutans binding activity to the hemorrhage area and binding activity to platelet activation.
The S. mutans were tested on rats. This was done to determine if S. mutans ability to bind to the collagen of a damaged cerebral blood vessel increasing hemorrhage area could occur in various organisms. The results were similar to that found in them mice. Thus, they were able to prove the enhanced hemorrhagic effects could occur in various organisms. While mice and rats are not humans, it does give evidence that S. mutans could cause similar effects in humans. The cnm proteins are thought to be the main issue. To see if the proteins themselves could cause excess hemorrhaging, recombinant pcnm proteins were administered to rats and the cerebral blood vessel was damaged. The results were similar to the cnm strains when 100 µg of recobinant cnm protein was injected into the blood stream. Further testing provided evidence showing that if the recombinant cnm proteins were administered prior to TW295, the TW295 microbes were blocked from binding the collagen by the recombinant proteins.
Conclusion
Research regarding the effects of S. mutans on both the dental and overall health of an individual has become more popular in recent years. As we begin to discover more about this pathogenic microbe, researchers have realized these microbes may be the reason for the well-known correlation between dental hygiene and cardiovascular disease. If these S. mutans do play as big as role in cardiovascular and hemorrhagic strokes as researcher hypothesize, then dentists and doctors could work together in preventing the harmful effects. This may be done by taking samples of the microbes within patients and screening for the cnm positive S. mutans strains. Doctors could possibly use a biological marker for the cnm gene or protein to screen patients. Then once the cnm positive strains are found, they could be killed using a drug that inhibits something unique within them. By using these screenings, doctors and dentists could in theory reduce cardiovascular disease and hemorrhagic stroke.
References
1. Loesche W.J. 1986. Role of Streptococcus mutans in Human Dental Decay. Microbiological Reviews 50(4):353-380
2. Centers for Disease Control and Prevention. 2001. Recommendations for Using Flouride to Prevent and Control Dental Caries in the United States. MMWR Recommendations and Reports 50(RR-14): 1-15
3. Shibata Y., Ozaki K., Seki M., Yamashita Y. 2003. Analysis of Loci Required for Determination of Serotype Antigenicity in Streptococcus mutans and Its Clinical Utilization. Journal of Clinical Microbiology 41(9): 4107-4112.
4. Ajdic D., McShan W.M., Mclaughlin R.E, Carson. M.B., Lai H., Ferretti J.J. 2002. Genome sequence of Streptococcus mutans UA159, a Cariogenic Dental Pathogen. Proceedings of the National Academy of Sciences. 99(22):14434-14439.
5.Maruyama F., Kobata M., Kurokawa K., Nishida K., Sakurai A., Nakagawa I. 2009. Comparative Genomic Analyses of Streptococcus mutans Provide Insights into Chromosomal Shuffling and Species-Specific Content. BMC Genomics 10:358-367.
6. Nakano K., Hokamura K., Taniguchi N., Ooshima T. 2011. The Collagen-Binding Protein of Streptococcus mutans is Involved in Haemorrhagic Stroke. Nature Communications 248(2): 1-10.
7. Aikawa C., Furukawa N., Watanabe T., Minegishi K., Furukawa A.2012. Complete Genome Sequence of the Serotype k Stretococcus mutans Strain LJ23. Journal of Bacteriology 194(10):2754-2759.
8.Northwestern Universituy IGEM 2013.
9. Loesche W.J. 2014. Chapter 99 Microbiology of Dental Decay and Periodontal Disease. Medical Microbiology 1-12.
10. Abranches J., Miller, J.H., Martinez P., Burne R.A., Lemos J.A. 2011. The Collagen-Binding Protein Cnm Is Required for Streptococcus mutans Adherence to and Intracellular Invasion of Human Coronary Artery Endothelial Cells. Infection and Immunity 79(6):2277-2288.
11. Zhao Q., Ikeda Y., Ihara H., Umemura K. 2003 Essential Role of Endogenous tissue Plasminogen Activator through matrix Metalloproteinase 9 Induction and Expression on Heparin-Produced Cerebral Hemorrhage afer Cerebral Ischemia in Mice. Blood Journal 103: 2610-2616.
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2009, Kenyon College.