The Effect of Acetylation and Deacetylation in Post-translational Regulation in Bacteria
Introduction
The processes of transcription and translation are extremely complicated and require a diverse range of enzymatic proteins and cellular machinery to create the desired result, protein. It seems that the simple conversion of DNA to mRNA in transcription and the transformation of mRNA to amino acids is straightforward and continuous. However, the complicated mechanisms involved in creating proteins are intricately regulated at many steps throughout the process.
Transcription begins with the recruitment of RNA polymerase by sigma factors, which recognize the promoters of specific genes to be transcribed. Sigma factors present a very coordinated form of regulation before transcription has even begun. Sigma factors recognize consensus sequences of DNA (common sequences amongst promoters) and therefore can initiate the transcription of multiple proteins (4). Thus, transcription is carefully regulated by the presence or absence of pertinent sigma factors, which is controlled by sensory mechanisms within the cell that determine stress levels and deficiencies of certain nutrients. The accumulation of a certain sigma factor can cause the transcription of a specific DNA sequence whose gene products are necessary in the cell at that particular time. After sigma binding, the DNA is unwound, and RNA polymerase binds tightly to the DNA. The RNA polymerase then moves along the sequence of DNA, creating mRNA. Transcription is terminated after the stop codon is reached. Even here, the presence of GC rich sequences regulates transcriptional speed and can temporarily halt DNA transcription to allow for the ribosome to catch up in translation. These halts in transcription, either via binding to the protein Rho or the formation of a GC-rich loop, can cause termination if the termination signals are met with RNA polymerase (4).
Before the RNA sequence can be bound to the ribosome and translated, the ribosome itself must come together. This is initiated with the transcription of rRNA genes that are processed by RNases and configured into a secondary structure. Simulaneously, the ribosomal subunits come together with the rRNA forming the ribosome. With the necessity of transcribing rRNA genes and the presence of the ribosomal subunits, regulation can control whether the cell can and wants to perform translation. Protein elongation occurs when mRNA at the ribosome is met with tRNAs carrying the appropriate amino acid that is coded by the mRNA. Even tRNAs require correct pairing with amino acids before reaching the ribosome; this process is performed by aminoacyl-tRNA transferases. This enzyme could also be modified and hindered, thus regulating translation. The completion of successful protein synthesis is not complete without precise folding of the protein. The correct folding of proteins is crucial to their functioning within the cell. Many proteins are modified by removing the N-formyl group from the N-terminus, by adenylylation, by phosphorylation, or by acetylation, thus regulating their function by modification of the protein itself (4).
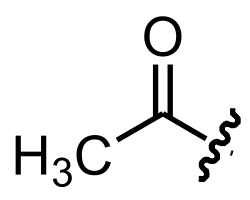
Regulation is occurring at frequent steps in this process of creating a functional protein. This regulation is of vital importance in most cells because of the colossal energy cost of transcription and translation. The formation and functioning of a ribosome alone can consume 40% of a bacterial cell’s energy, and therefore, the unnecessary production of proteins is extremely wasteful to a living cell (4). This limited energy must be conserved to carry out the numerous functions of the cell, and gene products must only be made when needed. Many stresses induce the need for certain gene products and suppress the need for others. For example, during starvation, many cells activate proteins that break down sugars for sustenance while others may activate ribosomal proteins that will transcribe necessary gene products. Other stresses, such as heat shock, intense pressure, and osmotic stress, may induce the formation or release of stability proteins to help the cell retain itself in extreme conditions. Stresses such as these can be sudden, and an immediate response may be necessary for the survival of the cell. The quickest method of regulation is post-translationally, or the modification of the proteins themselves. It is known that slight changes to the proteins can easily result in the fully inhibited functioning of the molecule. Therefore, as mentioned earlier, the simple addition of an acetyl group (acetylation) can inhibit the functioning of a protein or enzyme, thus controlling its ability to perform within the cell (Figure 1).
Acetylation and deacetylation was first determined in eukaryotes in the regulation of histones. Since, many other proteins have also been found to be acetylated, including proteins involved in DNA-protein interactions, transcription, and protein stability. Acetylation has also been found to have applications to age-related diseases, including cancer and cardiovascular diseases, for example (8). Eukaryotic histone proteins, which control chromatin, have also been found in Archaea. In 1990, histones homologous to eukaryotic histones were found in the methanogenic archaeon, Methanothermus fervidus. It has since been determined that histones are present in many members of the subdomain Euryarchaeota, but none have yet been found in Crenarchaeota. (9). The yeast Sir2 protein, an NAD+-dependent histone deacetylase, has also been found to have homologous Sirtuin proteins in archaeal, eubacterial, and human proteins (10). This indicates that there is a distinct evolutionary relationship between acetylation and deacetylation in the three domains and that the sirtuin proteins (nicotinamide adenine dinucleotide (NAD+)-dependent protein deacetylases) responsible for acetylation of histones in archaea and eukaryotes may have other targets in bacteria because bacteria have no histone encoding genes. In addition, there are four classes of eukaryotic sirtuins. It has been proposed that the first eukaryotic sirtuin was of class III and arose from an archaean parent, and class II sirtuins came from a probacterium parent. However, it was also found in the Kinetoplatida that a probacterium-derived sirtuin gene was present, indicating the possibility of horizontal gene transfer (11).
Here, the post-translational effect of acetylation and deacetylation on proteins in bacterial cells is explored, both in regulating proteins involved in cellular processes such as metabolism and in proteins involved in translation itself. In Salmonella enterica, an aerobic, gram-negative bacterium, acetylation of Acetyl-CoA Synthetase (Acs) inhibits its funtion, which transforms Acetate to Acetyl-CoA (6). A similar mechanism in the gram-positive soil bacterium Bacillus subtilis is used to regulate Acs by way of an operon (1). Lastly, Escherichia coli, the gram-negative bacterium commonly found in mammalial intestines, has shown to have numerous sites of acetylation in various proteins, including those directly involved in transcription (8). The regulatory effect of these modifications is of extreme necessity to cell survival because of its rapid response in changing conditions. The grave importance and prevalence of these acetylation and deacetylation systems makes this form of regulation an emerging area of study because of its application to industry, the environment, and plant, animal, and human pathogens.
Acetylation in Salmonella enterica
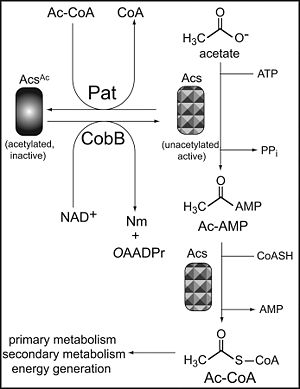
In the bacteria Salmonella enterica, post-translational modification of protein function has been determined to have a metabolic role by interacting with acetyl-coenzyme A (Ac-CoA) synthetase (Acs), the enzyme responsible for the conversion of acetate to the acetyl-adenosine monophosphate (Ac-AMP) intermediate and then to Ac-CoA (6). Acetyl-CoA is fundamental to cell metabolism, most importantly in the TCA cycle. Acetyl CoA is received mostly from the breakdown of pyruvate, but also from other catabolic pathways such as fat, amino acids, and benzoate breakdown. Acetyl CoA is taken up into the TCA cycle by incorporation via the combination of the acetyl group with oxaloacetyate. The cycle can then proceed to produce 3NADH, FADH2, and ATP, which then goes on to produce even more energy for the cell (4). Acetyl CoA is also very important to fermentation, where the acetyl group can be used to create Acetic acid, Isopropanol, Butanol, Butyric acid, or Ethanol (4). Thus, the conversion of acetate to Acetyl CoA is essential to cellular metabolism universally, so its regulation can induce a large effect on cells. However, the addition of an acetyl group to the residue lys609 of Acs blocks the conversion to the Ac-AMP intermediate (7). This acetylation, however, does not affect the formation of Ac-CoA from Ac-AMP (6). The acetyl donor necessary for acetylation to occur comes form Ac-CoA itself, therefore, a feedback inhibition mechanism.
In using this process to control Ac-CoA production and consequently energy production in the cell, Acs must be deacetylated to resume normal enzymatic function. The proteins that perform this function are called sirtuins, which are nicotinamide adenine dinucleotide (NAD+)-dependent protein deacetylases previously found in Eukaryotes and Archea to be important in chromosome stability and gene silencing (6). Specifically, however, in the deacetylation of Acs, it is the CobB protein (encoded by the cobB gene) that is responsible for the reactivation of the metabolic enzyme in S. enterica (7). CobB removes the inhibitory acetyl group from the inactive Acs, creating 2’-O-acetyl-ADPr, the functional protein (Figure 2) (6).
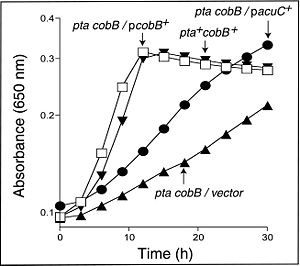
The gene encoding the protein acetyltransferase enzyme (Pat) responsible for the acetylation of Acs was recently identified in S. enterica. Pat was determined as the acetyltransferase enzyme by V. J. Starai and J. C. Escalante-Semerena who isolated strains of S. enterica lacking the putative Pat enzyme to determine the bacteria’s ability to grow at low acetate concentrations (<10mM). Tranposable elements were used to transform the cells and knockout different combinations of functions to determine their essential purpose in the regulation of cellular metabolism. At the low acetate concentration, four different strains were grown: wild type, pta cobB (lacking cobB sirtuin), pta pat (lacking pat enzyme), and pta cobB pat (lacking cobB sirtuin and pat enzyme). The pta pat and pta cobB pat strains grew at wild-type levels, and the growth of the pta cobB strain was greatly inhibited. This showed that the Pat enzyme was in fact responsible for the acetylation of Acs. This was evidenced because in each case when Pat was inactive, growth was normal, while growth was inhibited in the presence of Pat without the cobB sirtuin to reactivate enzymes, thus leaving them in their inactive states (6).
The molecular interactions involved in the binding of Pat to Acs have been found to occur at a lysyl residue, Lys609 (6). The residue Lys609 was identified by using a time course experiment of deacetylation of Acs. This method was based on the idea that if Acs was being acetylated at residue Lys609, the cobB sirtuin would be the deacetylation catalyst. The Lys609 binding site was confirmed with the use of Matrix-assisted laser desorption ionization—time of flight (MALDI-TOF) mass spectrometry peptide fingerprinting by looking for the number and locus of acetyl groups transferred from Pat. A high intensity signal was confirmed to represent the SGKAcIMR612 peptide in Acs where one acetyl group was observed (6). Later studies also determined Leu641, another residue lying close to the acetylation site, as a vital amino acid for the acetylation of Acs to occur. This was done by isolating unacetylated variants of Acs using a mutagenized plasmid and inhibiting acs genes (genes encoding acetyl-coenzyme A synthetase) and phosphotransacetylase genes. A single base substitution was induced in the acs gene resulting in the placement of a prolyl residue where the leucyl residue normally resides. Results showed that mutants were inviable with the addition of the propyl residue (7). Mutations were induced changing the leucine to Gln and Ala to check for specificity of the leucine residue. Results showed that Leu641 was necessary and was not specific to the propyl residue as initially tested (7).
This precise protein regulation of the vital metabolic enzyme acetyl-coenzyme A (Ac-CoA) synthetase shows a critical and well-directed function of acetylation in in Salmonella enterica.
Acetylation in Bacillus subtilis
In Bacillus subtilis, a similar post-translational modification of the enzymatic protein acetyl-coenzyme A synthetase, responsible for converting acetate to Ac-CoA, is involved, using the transfer of acetyl groups to inhibit enzymatic function. However, in B. subtilis bacteria, the catabolism of acetate requires the enzyme termed AcsA which is regulated by a three-gene operon, a unit of nucleotide sequences that share a single promoter and are transcribed together (1). The operon, acuABC (acetoin utilization) consists of two genes similar in function to the cobB and Pat acetylase/deacetylase system observed in S. enterica. The genes in the operon act on the substrate AcsA, where AcuA is a protein acetyltranferase and AcuC acts as a deacetylase. The third gene in the operon, AcuB, has an unknown function. In addition, while the cobB and Pat acetylase/deacetylase system observed in S. enterica is dependent on NAD+ for sirtuin activity, the activity of the AcsA enzyme does not require NAD+ (1).
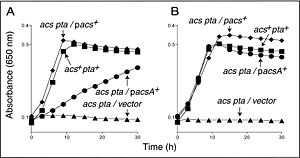
The functioning of these enzymes, AcuA and AcuC, was actually determined using strains of S. enterica whose use of Pat and CobB was suspected to be very similar to that in B. subtilis. By using S. enterica strains lacking both acs and cobB function, the effect of AcuA and AcuC could be determined by insertion into S. enterica cells via phage-mediated transduction. It was found that that AcuA restored the Acs function in S. enterica but was necessary to increase enzyme levels (Figure 3). AcuC restored some function in S. enterica but was not as efficient (Figure 4). Interestingly, very high levels of AcuA present in cells were found to be toxic, indicating that perhaps too many Acs proteins were acetylated, thus inhibiting function. To confirm the acetylation of AcsA by the AcuA protein, MALDI-TOF mass spectrometry was used, which showed that AcsA in B. subtilis was acetylated once at residue Lys549 (1).
Interestingly, the AcuA protein was found to have remarkable stability in stressful heat conditions. When B. subtilis was kept at 100°C for periods of 5, 30, and 60 minutes, 93%, 78%, and 15% of AcuA proteins remained intact and functioning (1). This is important for B. subtilis especially because if its presence in certain extreme mesophilic environments. Further experimentation had to deactivate enzymes with a high concentration of KCl salt because their incredible heat resistance prevented the complete inactivation of the enzyme with temperature control (2).
Apart from pure metabolic processes, the function of acuA has been correlated to the temporal gene expression during spore germination in B. subtilis. This is being investigated because during spore germination into the outgrowth phase, acuA was upregulated in the period between 80 and 100 minutes (2). This hints at the vast scope of processes that are regulated by acetylation and deacetylation in B. subtilis and in many other organisms.
Acetylation in Escherichia coli
Acetylation of lysine residues has diverse functions in Escherichia coli. Apart from the regulation of acetyl-CoA synthetase, CheY, which is responsible for chemotaxis, is regulated by means of acetylation by Acs (8). However, many acetylation sites in E. coli have been determined in proteins comprising various differing functions including carbohydrate metabolism, the TCA cycle, transcription, protein synthesis, and nucleotide synthesis. To determine the lysine-acetylated proteins found in E. coli, western blotting was performed using an anti-acetyllysine antibody to attach acetylated sites, which then shows up as a band when run on a gel. The presence of numerous bands indicated that many proteins had lysine-acetylated sites. Specifically, 125 acetylation sites were identified on 85 different proteins (8).
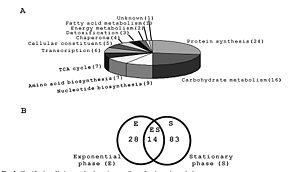
Lysine acetylation sites in E. coli can be broken down into the various functions that the proteins perform. Acetylation was observed in many metabolic enzymes (16) and enzymes involved in the TCA cyle (7). Acetylated enzymes include the glycolytic enzymes (phosphoglucose isomerase, glyceraldehyde 3-phosphate dehydrogenase (acetylated at 3 sites), dihydrolipoamide dehydrogenase, dihydrolipoamide acetyltransferase, enolase, phosphoglycerate kinase, phosphoglycerate mutase, and pyruvate dehydrogenase) and TCA cycle enzymes (citriate synthase, aconitate hydratase, citrate synthetase, isocitrate dehydrogenase (acetylated at 7 sites), dihydrolipoamide succinyltransferase, malate dehydrogenase, and succinyl-CoA synthetase. The vast number of enzymes listed here and found to be acetylated involving cellular metabolism indicate that the production of energy is most likely regulated by acetylation. Many proteins involved in amino acid biosynthesis (7), protein synthesis (24), and nucleic acid biosynthesis (9) were also observed (8) (Figure 5).
Aside from anabolism and catabolism, many proteins directly involved in DNA replication, stability, and transcription/translation were also found to be acetylated, indicating an even closer link to DNA regulation. In E. coli, 17 acetylated sites on 13 different ribosomal proteins were observed, meaning that acetylation may control the activity of ribosomes themselves. Among these acetylated sites, elongation factors (EF-T’s, G, and Tu), aspartyl-tRNA synthetase, and cysteome-tRNA ligase were all identified as lysine acetylated. Acetylation sites on other DNA-related proteins were also verified: transcription enzymes (RNA polymerase, topoisomerase I, ssDNA-binding protein, CRP transcriptional dual regulator, and Rho protein) and chaperones (GroEL, DnaK, GrpE, Hsp 70, and Hsp 90) (8). The considerable number of acetylated sites on proteins of various functions indicate that the significance of acetylation in bacteria is much broader that what is know currently about Acs and CheY, and its role may appear increasingly more important as further research in the area is conducted.
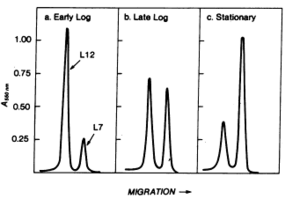
Lysine-acetylated sites found in this study were also investigated to determine a relationship between acetylation and growth phase. Acetylation sites did show a correlation with growth phase, where 15 sites were found in both exponential growth and stationary growth, 29 were found only in the exponential phase, and 82 were found only in the stationary phase. This indicates that acetylation is a likely player in the regulation of certain proteins involved in cellular growth. This was further shown when cells in stationary phase were transferred to a fresh LB medium and incubated for two hours. After this incubation period, only 2 sites were found to be lysine acetylated (elongation factor Tu and 50S ribosomal subunit protein) (8). An earlier study supports this result. The study looked at two ribosomal forms distinguished by differences in a protein in the 50S subunit, L7 (acetylated) and L12 (nonacetylated). Relative proportions of each of these forms in both exponential and stationary phases were measured using gel electrophoresis and a spectrometer. Results showed that during exponential phase, 85% of the area under the peaks on the spectra was under the nonacetylated L12 peak while in stationary phase, 65% was under the acetylated L7 peak (Figure 6) (3).
These results indicate that E. coli cellular proteins are most likely down-regulated by acetylation when cells are in stationary phase and are not rapidly growing and conducting metabolism, and acetyl groups are removed when exponential phase requires energy production and the synthesis of required proteins and genetic material. This has immense importance to cells encountering rapid changes in physiological conditions because dormant enzymes that are in their stable, acetylated state can be quickly revived and utilized, without requiring the long and energy taxing process of creating the proteins themselves. It is suspected that this acetylation may be controlled by intracellular levels of CoA and NAD+, which indicate the amount of energy and sustainance within the cell; however, this is still under investigation in E. coli and other bacteria (8).
Conclusion
The process of acetylation and deacetylation in bacteria is of extreme importance to their survival. Gene regulation by DNA folding and other transcriptional regulators has been known, but post-translational modifications of proteins serves as a means for cells to react quickly to changing environments and to simply convert from not producing energy to producing energy very quickly, for example. This mechanism is rapid because the proteins exist, needing only modification via a deacetylation enzyme or other enzyme based on the nature of the bound stabilizer. It seems that notably in bacteria, stabilizing regulation mechanisms are extremely important given the diverse range of environments in which these organisms have adapted to live. Adaptations, such as the use of stabilizing acetyl groups, provide the means for certain organisms to live in the truly remarkable environments found around the world today.
References
(1) Gardner, J. G., F. J. Grundy, T. M. Henkin, and J. C. Escalante-Semerena. August 2006. Control of Acetyl-Coenzyme A Synthetase (AcsA) Activity by Acetylation/Deacetylation without NAD+ Involvement in Bacillus subtilis. Journal of Bacteriology 188:5460-5468.
(2) Gardner, J. G. and J. C. Escalante-Semerena. July 2008. Biochemical and Mutational Analyses of AcuA, the Acetyltransferase Enzyme That Controls the Activity of the Acetyl Coenzyme A Synthetase (AcsA) in Bacillus subtilis. Journal of Bacteriology 190:5132-5136.
(3) Ramagopal, S. and A. R. Subramanian. May 1974. Alteration of the Acetylation Level of Ribosomal Protein L12 During Growth Cycle of Escherichia coli. Proc. Nat. Acad. Sci 71:2136-2140.
(4) Slonczewski, J. L., J. W. Foster, 2009, Microbiology: An Evolving Science: New York, W. W. Norton & Company, Inc. p. 257-300.
(5) Starai, V. J., H. Takahashi, J. D. Boeke, and J. C. Escalante-Semerena. 2004. A Link Between Transcription and Intermediary Metablism: A Role for Sir2 in the Control of Acetyl-Coenzyme A Synthetase. Current Opinion in Microbiology 7:115-119.
(6) Starai, V. J. and J. C. Escalante-Semerena. 2004. Identification of the Protein Acetyltransferase (Pat) Enzyme that Acetylates Acetyl-CoA Synthetase in Salmonella enterica. J. Mol. Biol 340:1005-1012.
(7) Starai, V. J., J. G. Gardner, and J. C. Escalante-Semerena. July 2005. Residue Leu-641 of Acetyl-CoA Synthetase is Critical for the Acetylation of Residue Lys-609 by the protein Acetyltransferase Enzyme of Salmonella enterica. The Journal of Biological Chemistry 280:26200-26205.
(8) Yu, B. J., J. A. Kim, J. H. Moon, S. E. Ryu, and J. Pan. January 2008. The Diversity of Lysine-Acetylated Proteins in Escherichia coli. J. Microbiol. Biotechnol. 18:1529-1536.
(9) White, M. F., S. D. Bell. 2002. Holding it together: chromatin in the Archaea. Trends in Genetics 18:621-626.
(10) Smith, J. S. et. al. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proceedings of the National Academy of Sciences of the United States of America 97: 6658-6663.
(11) Verdin, Eric. 2006.Histone Deacetylases. Humana Press 362.