The Microbiome of the Honeybee: Implications for Colony Health and Socialization
Introduction
The microbiome of the Honeybee plays a significant role in colony health by influencing various physiological processes and social behaviors.[1]. A core component of this microbiome consists of gut symbionts that affect not only individual health but also colony dynamics.[2] Honeybees' gut microbiota, such as Lactobacillus and Bifidobacterium, interact with their environment to promote colony hygiene, immune function, and nutritional health. For instance, substances like honey, royal jelly, and propolis, all of which have antimicrobial properties, help maintain the microbiome balance by inhibiting harmful pathogens while supporting beneficial microbes.[3]. The influence of the Honeybee microbiome extends beyond individual bees to the entire colony, impacting behaviors such as foraging, cleaning, and the communal care of larvae. The microbial communities present in the hive are crucial for fermenting and detoxifying pollen and nectar, thus ensuring the health of both the adult bees and their brood. These microbiomes are not only vital for protecting bees from pathogens but also aid in the transmission of beneficial microorganisms through the colony, contributing to the overall success of the hive.[4]. Honeybees are indispensable pollinators, supporting the growth of countless crops and maintaining biodiversity. Their microbiome, a community of microorganisms inhabiting their digestive tract, plays a vital role in their health, nutrition, and immunity, making it a key factor in the survival of these essential insects.[5]. This page explores the intricate relationship between Honeybee Microbiota, colony health and socialization, shedding light on the mechanisms that support bee resilience and the potential consequences of microbial imbalances for ecosystems and agriculture.
Genetics and Gut Bacteria
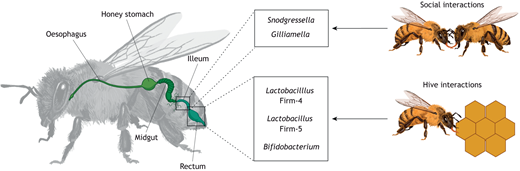
Honeybee gut microbiota, transmitted through social interactions[6], play a crucial role in supporting host health. While the microbial community remains relatively stable, individual variations and significant strain-level diversity have been observed among Honeybees. Although environmental factors influence the structure of the gut microbiota, the host genetics also shape the community. Given that bees within a colony are not genetically identical due to the queen's polyandry, it means that host genetics may contribute to shaping the microbiota structure.[7]. Gut communities are more different between genetically varied honey bees.[8].
Researchers have demonstrated that the composition of the honeybee gut microbiota is strongly influenced by host genetics.[9].
Genetically distinct bees exhibit significant differences in gut microbiota composition, both at the phylotype level and within specific microbial strains. For example, variations in the Snodgrassella alvi phylotype are shaped by host genotypes, with genetically diverse hosts preferentially selecting distinct microbial strains. Moreover, single-nucleotide polymorphism (SNP) patterns in Type IV pili loci differ significantly between strains from different hosts. Importantly, host genetics also influence specific microbiota functions; the abundance of a cluster of Bifidobacterium strains has been linked to variations in the host gluR-B gene, which encodes a glutamate receptor expressed in the bee brain. These interactions impact host neurobiology, as colonization by specific Bifidobacterium strains with polysaccharide utilization loci alters the alternative splicing of the gluR-B gene, leading to increased GABA levels in the brain. This highlights a complex interplay between host genetics, gut microbiota, and bee health.[10]
As in other animals, the bacteria in the gut of honey bees govern important functions critical to host health. There is a high degree of genetic diversity within each of the microbiome species. In the case of the gammaproteobacterial species Gilliamella apicola, it is shown that there's a link between genetic variation of isolates and functional differences suggesting that niche partitioning within this species emerged during evolution with its bee hosts.[11]
The consistent presence of only a few species, combined with strain variation within each of these species, makes the gut microbiota of social bees an ideal model for studying functional, structural, and evolutionary aspects of host-associated microbial communities: many characteristics resemble the gut microbiota of humans and other mammals, but the complexity is considerably reduced.[12]
Gut Microbiome and Honeybee Health
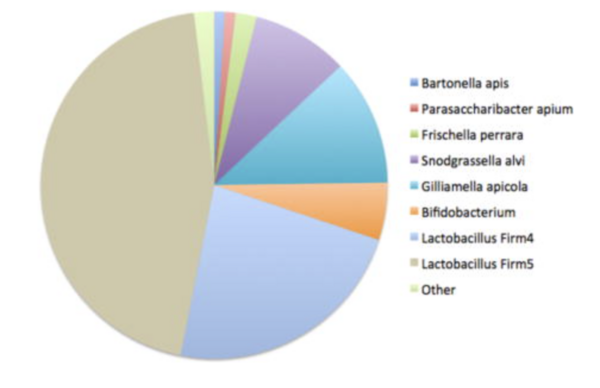
The honeybee gut hosts a simple, conserved microbiome comprising core bacterial species. These microbes aid in digestion, immune defense, and detoxification of harmful compounds[13]. Only five species’ clusters form the core of honey bees’ gut microbiome: Snodgrassella alvi, Gilliamella apicola, Lactobacillus Firm-4, Lactobacillus Firm-5, Bifidobacterium asteroides. Less numerous groups are: Frischella perrara, Bartonella apis, Bombella apis and Commensalibacter sp[14].
Perturbance of the Honeybee core microbiome through pesticide, antibiotic or experimental manipulation causes numerous physiological impacts, ranging from increasing disease susceptibility[15] to delaying behavioral development[16]. Thus, honeybee gut symbionts are essential to the proper functioning of Honeybee physiology, including their metabolism, immune system function[17] and developmental processes such as weight gain by stimulating host pathways that synthesize key hormones such as juvenile hormone and vitellogenin[18]. Additionally, Honeybee workers exhibit a diverse array of behaviors that change with age, allowing them to organize within their labor castes in both time and space[19]. This organization is influenced by environmental and social factors, ensuring the colony thrives across different seasons[20]. While it is well understood that hormones play a significant role in regulating these seasonally shifting task specializations, the gut microbiome also is a key factor in this process. Recent research[21] has shown that the abundance of core gut symbionts varies with the seasons. As a result, Honeybee behavior is shaped by the interplay between their physiological traits, environmental conditions, and gut microbiota. Although the core gut microbiome of Honeybees is generally stable, the differences in the abundance of specific gut symbionts among worker behavioral castes may influence their division of labor[22].
Studies on core bee gut species, such as Gilliamella apicola, Lactobacillus species, and Bifidobacterium, reveal their ability to digest and metabolize a wide variety of plant-derived carbohydrates. Comparisons between microbiota-free bees and those with a conventional gut microbiome demonstrate significant physiological impacts of the microbiome. These include increased gut size, greater weight gain after eclosure, enhanced insulin and vitellogenin signaling, and improved sucrose sensitivity. These physiological traits are closely linked to bee health, immune function, and resistance to stress. Additionally, the gut microbiome significantly influences the composition of short-chain fatty acids in both the gut and hemolymph; for instance, butyrate is prevalent in conventional bees but entirely absent in microbiota-free bees.
Influence of Gut Bacteria on Honeybee Castes
Nurses
The youngest worker caste, known as nurses, have notably higher levels of Lactobacillus Firm-4, Firm-5, and Bifidobacterium compared to foragers, the oldest worker caste[23]. Since Lactobacillus and Bifidobacterium are crucial for breaking down pollen, an abundance of these microbes supports the nurses' role in feeding and nurturing the brood[24]. Nurses consume pollen, which stimulates the development of their hypopharyngeal glands to produce food for larvae[25]. Furthermore, the colonization of Gilliamella in the ileum may play a key role in preparing nurses for their future role as foragers, helping them cope with the significant oxidative stress associated with foraging. For instance, the inability of Gilliamella apicola to colonize in nurses is linked to reduced nutrient metabolism, potentially leading to premature transitions into foragers with diminished foraging capabilities.
Foragers
Foragers represent a well-studied behavioral caste of Honeybees. A crucial factor in their behavioral development is sugar sensitivity, which impacts their olfactory learning, memory, and foraging preferences[26]. Notably, nectar foragers exhibit lower sucrose sensitivity compared to pollen foragers, enabling them to identify highly rewarding nectar sources in their environment[27]. This sugar sensitivity is governed by the insulin-like signaling pathway in the honeybee brain[27]. Evidence suggests that gut microbes play a role in modulating sucrose sensitivity by promoting insulin production, which significantly influences forager behavior[28]. Specifically, the insulin-like peptide genes ilp1 and ilp2 are less active in bees with depleted gut microbiota, and these bees also show reduced responsiveness to sugar rewards.
Queens
Understanding the regulation of Honeybee societies requires considering their reproductive members. In these eusocial colonies, a single queen produces workers, while a small number of drones handle genetic dissemination during the summer[29]. The gut microbiome of queens is notably distinct from that of workers. In young queens, enteric bacteria such as Escherichia and Gilliamella dominate the gut, but by 14 days of age, the gut microbiome shifts to being dominated by Parasaccharibacter apium and related Alphaproteobacteria strains associated with worker[30] hypopharyngeal glands which may be interesting as it might contribute to her egg-laying capabilities by providing the energy needed for such costly labor[31]. This change is likely linked to the queen's lifelong diet of royal jelly, which is produced by worker hypopharyngeal glands and may influence the composition of the queen's gut microbiome.
Drones
The drone gut microbiome remains the least explored among Honeybee castes, likely due to their seemingly limited role in the colony and the focus on the more behaviorally diverse worker bees. Drones' guts are primarily dominated by Lactobacillus Firm-4 and Firm-5 species[32]. Emerging research suggests a link between gut microbiome health and spermatogenesis[33], indicating that the gut microbiome could impact drone reproductive success by influencing sperm quality and supporting the energy demands of mating flights.
Conclusion
The microbiome of the honeybee plays an integral role in colony health and socialization, shaping the physiological and behavioral traits critical to the survival and success of these eusocial insects. From nutrient metabolism and immune defense to the regulation of caste-specific roles and reproductive functions, the gut microbiota constantly influences individual and colony dynamics. These intricate interactions highlight the microbiome's pivotal role in fostering resilience and adaptability within honeybee societies. Understanding these relationships is essential for safeguarding honeybee populations, which are vital to biodiversity and global agriculture and it also paves way for studying the relationships between behavior and gut bacteria in a wide range of organisms.
References
- ↑ https://pmc.ncbi.nlm.nih.gov/articles/PMC3555888/pdf/gmic-4-60.pdf
- ↑ https://www.science.org/doi/full/10.1126/sciadv.abd3431
- ↑ https://pmc.ncbi.nlm.nih.gov/articles/PMC8272120/
- ↑ https://www.jstor.org/stable/pdf/32825.pdf
- ↑ https://academic.oup.com/femsre/article/37/5/699/542120
- ↑ https://www.sciencedirect.com/science/article/pii/S221112472400737X
- ↑ https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-021-01174-y/figures/1
- ↑ https://journals.plos.org/plosone/article/figure?id=10.1371/journal.pone.0036393.g001
- ↑ https://journals.plos.org/plosone/article/figure?id=10.1371/journal.pone.0036393.t002
- ↑ https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-021-01174-y
- ↑ https://pmc.ncbi.nlm.nih.gov/articles/PMC3390884/
- ↑ https://www.cell.com/cell/fulltext/S0092-8674(12)00104-3
- ↑ https://www.sciencedirect.com/science/article/pii/S2214574517301761
- ↑ https://pmc.ncbi.nlm.nih.gov/articles/PMC5648345/
- ↑ https://www.pnas.org/doi/abs/10.1073/pnas.1803880115
- ↑ https://journals.biologists.com/bio/article/9/11/bio053884/225338/Antibiotics-in-hives-and-their-effects-on-honey
- ↑ https://journals.biologists.com/bio/article/9/11/bio053884/225338/Antibiotics-in-hives-and-their-effects-on-honey
- ↑ https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.2003467
- ↑ https://link.springer.com/article/10.1007/BF00299306
- ↑ https://www.sciencedirect.com/science/article/pii/S0022519301924270
- ↑ https://academic.oup.com/ismej/article/14/3/801/7474760
- ↑ https://link.springer.com/article/10.1007/S00040-018-0624-9
- ↑ https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0123911
- ↑ https://enviromicro-journals.onlinelibrary.wiley.com/doi/full/10.1111/1462-2920.12526
- ↑ https://royalsocietypublishing.org/doi/full/10.1098/rspb.2017.2849
- ↑ https://www.pnas.org/doi/epdf/10.1073/pnas.0800630105
- ↑ 27.0 27.1 https://link.springer.com/article/10.1007/s00265-019-2644-5
- ↑ https://www.mdpi.com/2075-4450/3/4/1084
- ↑ https://www.pnas.org/doi/abs/10.1073/pnas.0505858102
- ↑ https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0200527
- ↑ https://www.sciencedirect.com/science/article/pii/S0016508514002194
- ↑ https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0123911
- ↑ https://gut.bmj.com/content/69/9/1608.abstract
Edited by [Kudzaishe Mhere], student of Joan Slonczewski for BIOL 116, 2024, Kenyon College.