Thiocapsa roseopersicina
Classification
Bacteria › Proteobacteria › Gammaproteobacteria › Chromatiales › Chromatiaceae › Thiocapsa
Species
Thiocapsa roseopersicina
Synonyms: Thiocapsa minima, Thiocapsa floridana
|
NCBI: Taxonomy |
Description and Significance
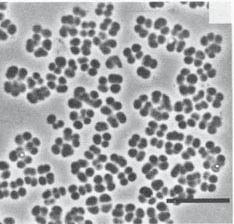
Thiocapsa roseopersicina was discovered by Sergei Winogradsky in the 1880s when he was studying sulfur bacteria. (9) Thiocapsa roseopersicina is a purple sulfur bacteria from the family of Chromatiaceae. (4) It is gram negative and non-motile. The bacterium is either rose colored or milky white based on the growth conditions. (1)
Thiocapsa roseopersicina has optimal growth in a medium with 0.3 M sodium chloride. This is a lower concentration than that of the seawater that T. roseopersicina is found, but that is common for marine/halotolerant organisms to have different optimal conditions than that of the environment they are isolated from. This is a good survival technique as the osmolarity of the marine water fluctuates from various occurrences like rain or evaporation. (5)
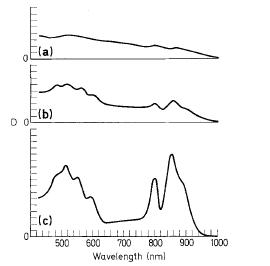
The bacterium is mainly characterized as an anoxygenic photosynthetic bacteria but it also has chemolithotropic capabilities that are dependent on the environment. Bacteriochlorophyll a is the main light harvesting pigment. The peak optical absorbance is between 800-930 nanometers which is near infrared range. The rose color of the bacteria is formed by the optical activity of the carotenoids (pigments of chloroplast). Spirilloxanthin is the most abundant carotenoid in Thiocapsa roseopersicina. (2)
Thiocapsa roseopersicina can fix atmospheric N2. This process is usually accompanied by H2 production. T. roseopersicina has three different Nickel Iron hydrogenases (HynSl, HupSl, HoxEFUYH). (8) Hydrogenases catalyze the reaction: 2H+ 2e ↔ H2. This reaction is reversible and catalyzes H2 generation and H2 oxidation. H2 oxidation is important for fuel cells and research is being performed on these hydrogenases as they generate a potential energy carriers. The hydrogenases of T. roseopersicina are particularly important in that they are tolerant to oxygen. Most hydrogenases are not oxygen tolerant, and those that are favor catalyzing the reaction taking up H2 instead of producing it. (7)
Genome Structure
The genome is currently under construction. The genome of Thiocapsa roseopersicina was sequenced in order to help understand the bacterium’s hydrogen metabolism and related metabolic pathways. Shotgun sequencing was done and the results were assembled into 2400 contigs. Currently, the sequencing of 3000 cosmid clones are in progress in order to link the previous contigs into larger contigs as well as fill in gaps between the contigs. Despite the unfinished assembly, many genes were discovered in preliminary results Thiocapsa roseopersicina genome assembly. (8)
One of the initial findings was the Tat gene sequence in the Thiocapsa roseopersicina genome. The Tat pathway is the twin-arginin translocation pathway. It is the pathway of transporting membrane-associated hydrogenases to the periplasmic space. Another discovery from the shotgun genome library is that the Hyn hydrogenase genes have two extra open reading frames. They are named isp1 and isp2 and are located in the middle of the genes that code for the small and large hydrogenase subunit. (8)
Cell Metabolism and Structure
Thiocapsa roseopersicina has spherical cells that can form tetrads. The cells are 1-3µm in size. The cells do not have gas vesicles. The guanine and cytosine content is 65.3%. T. roseopersicina can use hydrogen, sulfide, thiosulfate, acetate, propionate, pyruvate, malate, succinate, fumarate, fructose, or glycerol as substrates. It cannot use formate or glucose as substrates though. (3)
Thiocapsa roseopersicina has two different types of metabolism that it can use based on the environment. Its main metabolism is anaerobic photosynthesis which uses reduced sulfur compounds as electron donors. The secondary metabolism is chemolithotrophy in dark environments, and it uses sulfide or thiosulfate as electron donor and oxygen as the terminal electron acceptor. The discovery of the secondary metabolism of chemolithotrophy was supported by the presence of the active ribulose biphosphate carboxylase enzyme which shows that carbon dioxide is assimilated using the reductive pentose pathway. (1)
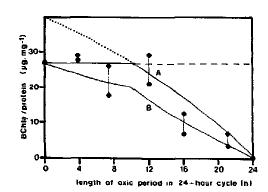
The metabolism changes based on the ecosystem conditions. The marine environment in which the bacterium is found has a steep gradient of oxygen and sulfur. When the bacterium is in the presence of oxygen, the bacteriochlorophyll a and the carotenoid synthesis is inhibited. This makes it harder for T. roseopersicina to use its photosynthetic metabolism, so it starts to shift to a chemolithotrophic metabolism. When the T. roseopersicina is growing phototrophically, the color of the bacteria is a bright rose. When the bacterium shifts completely to a chemolithotropic metabolism, the color shifts to a milky white. This is caused by a washout of the bacteriochlorophyll with increasing time in an oxygenic environment. (1)
Thiocapsa roseopersicina grows best when it can use both metabolisms simultaneously. This usually occurs in microaerobic conditions of 0-20µM of O2. Photosynthesis is preferred under electron donor limiting conditions, especially when the cells have a lot of pigment like the bacteriochlorophyll a and the carotenoids. When T. roseopersicina is growing under chemolithotropic conditions, it has to compete with colorless sulfur bacteria. (1) Therefore, when T. roseopersicina is using its photosynthetic metabolism, it has a faster growing rate. The bacterium reaches a stationary phase at 3-4 days with the photosynthetic conditions whereas it reaches a stationary phase at 7-12 days with the chemolithotrophic conditions. (6)
Ecology
Thiocapsa roseopersicina is mainly found in microbial mats in hypersaline and marine environments. Purple sulfur bacteria like T. roseopersicina are limited to a narrow zone vertically that is 1-2 millimeters thick. This area usually fulfills the purple sulfur bacteria’s need for light and sulfide. This area also has opposing gradients of sulfide and oxygen which fluctuates based on the day and night cycle. T. roseopersicina’s interchangeable metabolism is useful for surviving the oscillating conditions. (1)
Thiocapsa roseopersicina has often been found in anaerobic waste stabilization ponds that have a lot of dissolved organic matter. It is often the dominant anoxygenic phototrophic bacterium in microbial mats, which may be in part because of its flexible metabolism. Thiocapsa roseopersicina can form a visible pink to purple patch in the water or on the sediment that it is growing upon. (2)
T. roseopersicina coexists with M. chtonoplastes and other cyanobacteria in microbial mats. These cyanobacteria oxidize sulfide to thiosulfate. T. roseopersicina uses the thiosulfate, especially in their chemolithotrophic metabolism. (1)
Application to Biotechnology
Thiocapsa roseopersicina is an important bacterium for microbial detoxification. It is one of the few bacteria that can completely oxidize dimethyl sulfide (DMS) to sulfate. The process starts with the presence of dimethylsulfoniopropionate (DMSP) in the environment. T. roseopersicina can cleave DMSP to DMS and acrylate under anoxic, light conditions. DMS and acrylate inhibit microbial growth at high concentrations. DMS in particular also contributes to acid precipitation and has a role in the global sulfur cycle. Under oxic, light conditions, T. roseopersicina can convert DMS to sulfate as well as convert acrylate to propionate. The main drawback is that not all of the DMS is converted to sulfate so there is still a significant amount of DMS in the environment. (5)
The Craig Ventor Institute has used Thiocapsa roseopersicina as a vector for testing a novel hydrogenase gene from a microbial metagenomics dataset. This method is a culture independent technique to assess novel genes from microbial metagenomics data. Previously, only hydrogenases from cultivable microbes were screened. Metagenomics datasets often lead to the discovery of new genes from uncultivable or unknown organisms. With this method, hydrogenase genes from uncultivable or unknown bacteria can be expressed by T. roseopersicina with the use of the bacterium’s hydrogenase accessory genes. This was the first instance where a functional hydrogenase was converted from environmental DNA. (7)
T. roseopersicina can also be used to monitor bacteriochlorophyll a-containing cells with optical instruments. The optical characteristics of the bacterium were assessed. These characteristics will be used for developing a technique that will be able to use optical instruments to quantify photosynthetic bacteria. This technique will be especially useful in wastewater treatment plants as it can be used to remotely monitor bacteriochlorophyll a-containing cells either continuously or periodically.(2)
References
(4) Imhoff, Johannes F. Prokaryotes. Vol. 6. New York: Springer New York, 2006. 849-861.
Author
Page authored by Tasneem Pierce and Nirzari Pandya, student of Professor Jay Lennon at Michigan State University.