User:S4392251
Jocelyn Xanthopoulo, Bench B, 43922510, 31/08/2016 [1]
Porphyromonas gingivalis
Classification
Higher order taxa
Bacteria; Fibrobactere-Chlorobi-Bacteroidetes superphylum; Bacteroidetes; Bacteroidales; Porphyromonadaceae; Porphyromonas[1]
Species
Species name: Porphyromonas gingivalis[2]
Type Strain: W83[2]
Description and Significance
Porphyromonas gingivalis is a black pigmented, gram negative, non-motile rod shaped bacterium, found predominantly in the subgingival plaque of individuals with periodontal disease[3]. Strain W83 was initially isolated in the 1950’s by H. Werner in Bonn, Germany from an undocumented infection of the human oral cavity[2]. P. gingivalis has been successfully cultured on Wilkins-Chalgren medium or on horse blood agar plates with added hemin and vitamin K1, for 48 to 72 hours in anaerobic conditions at 37°C, forming shiny black colonies due to haem accumulation[4][5]. P. gingivalis is a clinically relevant bacterium that has frequently been isolated in the presence of several oral diseases, including oral abscesses, dental caries, pulpal infections, and most importantly periodontitis[3]. Periodontal disease encapsulates a multitude of oral infections of differing severity, which can present as mild gingival inflammation or chronic destruction of connective tissues and bone, ultimately resulting in tooth loss described as periodontitis[6]. Presence of P. gingivalis in the subgingival plaque has been consistently linked to chronic and severe periodontitis due to its associated virulence factors[6]. Continued study of P. gingivalis is vital as an estimated 10-15% of the world’s population is expected to suffer from periodontitis[7]. P. gingivalis presence has also been associated with systemic conditions including, rheumatoid arthritis, coronary artery disease and preterm birth[8].
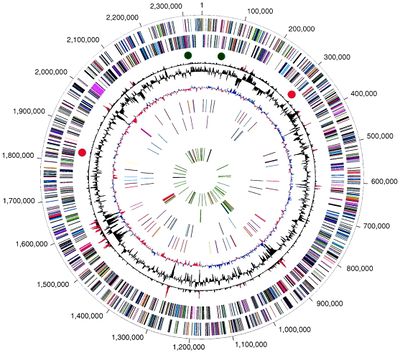
Genome Structure
The W83 strain of P. gingivalis has a circular DNA genome that is 2343479 base pairs long, with 1,990 open reading frames (ORFs) and 1909 protein coding genes identified using whole genome sequencing[1]. The GC content of the genome is 48.3% (Figure 1)[1]. The genome also contains 65 RNA genes, including two structural RNA genes, four ribosomal operons, and 53 tRNA genes that give P. gingivalis specificity for all amino acids[1]. An estimated 6% of the genome is repetitive elements, including transposable elements and DNA repeats such as direct repeats and clustered regularly interspaced short palindromic repeats (CRISPRs)[1]. P. gingivalis is present in the oral cavity with a multitude of other microorganisms capable of undergoing frequent and complex interactions, which enable the community to rapidly respond to environmental changes[9]. In turn P. gingivalis is capable of frequent DNA acquisition events through horizontal gene transfer, to give it substantial adaptive potential[9]. Genes distinct to the virulent W83 strain of P. gingivalis, which separate it from avirulent P. gingivalis strains and other oral microbiota are Arg-gingipain (Rgp) and Lys-gingipain (Kgp), encoding proteases, and Mfa1 and FimA genes, encoding the major and minor fimbriae respectively[10].
Cell structure and metabolism
Cell Wall and Metabolism
P. gingivalis has a gram-negative cell wall composed mainly of lysine rich peptidoglycan layers[1]. Iron acquisition is vital for P. gingivalis growth, as such this bacterium must have the capability of sequestering iron from the host. P. gingivalis does not have the typical siderophores in its cell wall to acquire iron, but instead uses outer membrane receptor proteases, lipoproteins, and gingipain proteases[10]. Haem accumulation causes the black pigment seen during culture of P. gingivalis[10]. Gingipain proteases also degrade host serum albumin to provide a source of nitrogen and carbon[10]. P. gingivalis is a facultative anaerobe, as it preferentially colonizes the subgingival sulcus, an anoxic and low sugar environment[1]. Whole-genome analysis (WGA) of P. gingivalis provides evidence that it can metabolize particular sugars, including melibiose, galactose, starch, and maltodextrin[1]. WGA also shows evidence of complete pathways for the degradation of complex amino sugars, whether these sugars are utilized for energy is unclear[1]. However it is known that at least 11 amino acids can serve as substrates for energy production, the amino acids are sourced from degraded host tissue[1]. Overall P. gingivalis is a very poor metabolizer of organic nutrients and carbohydrates in general[1]. Major fermentation products produced based on WGA are propionate, butyrate, isobutyrate, isovalerate, acetate, ethanol, and butanol, many of which are toxic to human host tissues[1].
Biofilm Formation
P. gingivalis is a late colonizer of multispecies subgingival plaque biofilms. Attachment is mediated by interaction between bacterial major fimbriae and surface receptors of intermediate colonizers[11]. According to the model of periodontopathic biofilm formation early colonizers include Streptococci and Actinomyces species, with intermediate colonizers including Fusobacterium nucleatum, which attract late colonizers usually including pathogenic gram-negative bacteria[11]. Alteration of the subgingival microenvironment with the colonization of each new species allows the attachment and colonization of the next stage of bacteria, through reduction of oxygen, and providing growth substrates used by the next species. Within the biofilm environment P. gingivalis undergoes complex interactions with other bacteria present using quorum sensing. Interaction occurs mainly between P. gingivalis and other late stage colonizers in periodontitis patients, including Treponema denticola and Tannerella forsythia[12]. P. gingivalis has been shown to influence the other microbiota present in the biofilm. Evidence suggests that communication of P. gingivalis with other bacterial microbiota is crucial for pathogenicity and disruption of host immune response[12]. Additionally P. gingivalis and T. denticola display synergy in biofilm environment, enhancing the pathogenesis of the subgingival plaque[12].
Ecology
P. gingivalis typically colonises the subgingival sulcus of the oral cavity and is an obligate anaerobe[9]. P. gingivalis is able to undergo fermentation of amino acids to produce energy, giving it the ability to survive deep in the periodontal pocket where sugar and oxygen availability is low[9]. P. gingivalis is an opportunistic pathogen, and is found as part of the indigenous oral microbiome in healthy individuals[9]. Other human environmental niches P. gingivalis has been found to colonise include the respiratory tract, upper gastrointestinal tract, colon, and in rare cases the vagina to cause bacterial vaginosis[6]. P. gingivalis is a secondary or late coloniser of subgingival plaque biofilm environments, adhering to early colonizers such as Streptococci and Actinomyces species[11]. Disease is observed when the subgingival biofilm begins to be composed primarily of secondary colonisers, usually pathogenic gram-negative bacteria, including P. gingivalis[11]. However periodontitis can be easily prevented by mechanically removing the plaque biofilm, usually using a toothbrush[11].
Alteration of the composition of the subgingival biofilm leading to disease presentation is facilitated by the host immune response[11]. Primary colonisers accumulate in the subgingival sulcus, forming a biofilm which induces a neutrophil mediated immune response, causing inflammation and redness of the gingiva, clinically referred to as gingivitis[11]. Progression from gingivitis to periodontitis occurs when an altered immune response is undertaken by the host, specifically a lymphocytic and plasma cell mediated immune response[11]. This immune response causes cell destruction, causing further inflammation and bleeding. Leading to modification of the subgingival environment altering the composition of the biofilm to better accommodate gram-negative pathogenic bacteria[12].
Pathology
P. gingivalis has been isolated from 85.75% of subgingival plaque samples from individuals with chronic periodontitis[13]. However, other bacteria have been implicated in the destruction of periodontal tissue leading to chronic periodontitis in the absence of P. gingivalis[13]. The ability of P. gingivalis to colonise and invade host cells of the subgingival sulcus is determined by the virulence factors encoded by the Arg-gingipain (Rgp), Lys-gingipain (Kgp), FimA and mfa-1 genes[10].
Gingipain Genes
Rgp is vital for nutrient acquisition through the degradation of large host proteins to provide the bacterium with nitrogen, carbon, and iron sources[10]. Rgp is also necessary for adhesion and invasion of host cells, as well as processing the precursor proteins of major fimbriae, vital for attachment to epithelial surfaces[10]. Similarly Kgp is involved in host colonisation, conferring the ability to bind matrix proteins fibrinogen and fibronectin[10]. The Kgp gene also confers the ability for P. gingivalis to avoid opsonisation and subsequent phagocytosis by host leukocytes through cleavage of IgG and opsins C3b and iC3b[10]. Together the gingipain genes cause the tissue damage symptoms associated with periodontitis, including degradation of matrix proteins, collagen and fibronectin[10]. Leading to decreased interaction between host cells and extracellular matrix causing destruction of periodontal tissues[10]. Gingipain genes also interfere with the host immune response by cleaving antibodies and pro-inflammatory cytokines, specifically IL-8, as well as chemokines to delay neutrophil recruitment[10]. Reducing bacterial clearance, allowing further P. gingivalis colonisation to disrupt the host-microbial homeostasis, leading to tissue and bone loss.
Fimbriae
The FimA and mfa-1 genes encodes the major and minor fimbriae respectively, which have the ability to bind extracellular matrix proteins, salivary enzymes, and epithelial cell α5β1-integrin and primary colonising bacteria[14]. Fimbriae are critical for P. gingivalis virulence and survival through contribution to cellular attachment, colonization and subversion of host innate immune responses[14].
Polysaccharide Capsule
The composition of the polysaccharide capsule of P. gingivalis is strain dependent, with sugar composition being the main point of difference[6]. P. gingivalis displays approximately six serotypes of capsular antigens, K1–K6. Highly encapsulated strains of P. gingivalis show decreased autoagglutination, resistance to phagocytosis, and have increased hydrophilicity than non-encapsulated strains[6]. Evidence has shown that the capsule of P. gingivalis assists in facilitating adhesion to host epithelial cells[6]. Alternatively the capsule also assists in co-aggregation of P. gingivalis and Fusobacterium nucleatum, another pathogen associated with periodontitis[6].
Lipopolysaccharide (LPS)
Lipopolysaccharide is a part of the bacterial outer membrane, it consists of a polysaccharide or O-antigen, a non-repetitive oligosaccharide core and a hydrophobic lipid A or endotoxin[6]. The lipid A is the portion of the LPS molecule that is biologically active which interferes with the host’s innate immune system by interacting with cell toll-like receptors 2 and 4b[6]. To affect host immune signaling and reducing bacterial clearance from the host. LPS is also plays a vital role in the structural integrity of gram-negative bacteria[6].
Application to Biotechnology
P. gingivalis has been associated with serious human disease, and as such is classified as a human pathogen. Subsequently biotechnological efforts focus on vaccine production. Ross et al. (2001) identified 120 potential vaccine candidate genes using a combination of whole genome sequencing and bioinformatic methods[15]. Two of the ensuing recombinant proteins, PG32 and PG33, induced a protective immune response in the murine model[15]. Further research conducted by Yonezawa et al. (2001) identified the Rgp-gingipain genes RgpA and RgpB as possible vaccine targets[16]. DNA RgpA vaccines induced high levels of serum antibodies against P. gingivalis, causing a reduction of the proteolytic activity of RgpA and RgpB and inhibited the binding mediated by these genes[16]. The results from both of these studies suggest that vaccines can induce protective host antibody responses against P. gingivalis and could confer protective immunity against future P. gingivalis infection[15][16]. Similarly effort has been put into using the Rgp-gingipain and Kgp-gingipain protease gene products as drug targets for use once an individual has presented with gingivitis, as a prevention method, or with mild to severe periodontitis as a treatment method[10][17].
Current Research
Currently research is continuing into the complex interactions P. gingivalis undergoes in the subgingival biofilm environment[12]. Specifically into the relationship between P. gingivalis and T. denticola in the late stage biofilm present in individuals with periodontitis[12]. The presence of P. gingivalis mediates the attachment of T. denticola, and evidence from flow cell studies implies that these bacteria act together to create a synergistic effect on disease progression and biofilm pathogenesis[12]. Other current investigations are occurring into possible treatment targets on P. gingivalis, specifically the use of plant derived compounds as therapeutic agents[15]. These are appealing as treatment options as they show less toxicity toward host cells[15]. Investigations into treatments targeting vital virulence factors is also occurring, specifically into the gingipain proteases, Arg-gingipain (Rgp) and Lys-gingipain (Kgp), as they are necessary for iron acquisition[10]. Without high levels of iron P. gingivalis is unable to grow and divide correctly, reducing its pathogenic effects on the host[10].
References
- ↑ MICR3004
This page is written by Jocelyn Xanthopoulo for the MICR3004 course, Semester 2, 2016
