User:S4442692
Simone Webb Bench B 02/09/2016 [1]
Classification
Higher order taxa
Bacteria – Bacteria – Bacteroidetes – Flavobacteria – Flavobacteriaceae – Flavobacteriaceae – Capnocytophaga
Species
Capnocytophaga gingivalis . Type strain is 27 [1].
Description and significance
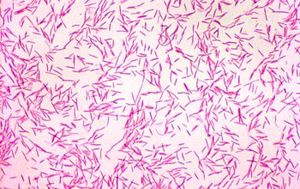
Capnocytophaga gingivalis is a gram negative, mesophilic and non-sporulating rod-shaped bacteria implicated in opportunistic periodontal disease [1, 2]. In 1979, Leadbetter et al proposed the genus ‘Capnocytophaga’, to include three morphologically and physiologically distinct species: Capnocytophaga ochracea, Capnocytophaga gingivalis and Capnocytophaga sputigena. The Capnocytophaga are gliding bacteria which constituted a predominant part of the cultivatable microbiota isolated from the gum in the Leadbetter et al study. The ability to attack polysaccharides and their capnophilic CO2-dependent growth (optimum growth in enrichment of carbon dioxide at 5-10%) influenced the naming of ‘Capnocytophaga’. ‘Gingivalis’ further refers to the fact that C. gingivalis are found on gingival crevices in the human oral cavity. C. gingivalis is clinically significant due to its ability to systemically spread from biofilm in the human oral cavity to the blood, where it is most recently implicated in COPD and bacteraemia [3, 4].
Genome structure
The ATCC 33624 strain of Capnocytophaga gingivalis was shotgun sequenced and submitted to NCBI as a reference for the Human Microbiome Project in 2009 [5, 6]. ATCC 33624 has a linear 2.67 Mb genome which consists of 2,507 genes coding for: 2,364 proteins, 8 rRNA’S, 47 tRNA’s, 1 ‘other’ RNA and 87 pseudogenes. The genome also consists of antibiotic-resistance plasmids which can be horizontally transferred to members of the same genus [4, 7].
Cell structure and metabolism
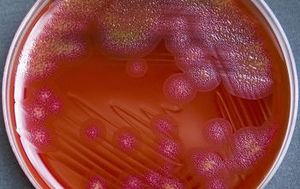
C. gingivalis are gram negative, short, rod-shaped cells, with a size of around 0.42-0.6 by 2.5 - 5.7 µm [1]. In early microscopic studies, the cell surface was observed to be devoid of fimbriae, flagella, and pili [8]. Colonies of C. gingivalis are flat, thin, slightly yellow/pink and were seen by Leadbetter at al to have ‘uneven edges’ with ‘finger-like’ projections. Colonies of C. gingivalis were further recorded to spread far, even centimetres, away from the initial inoculation site on the agar media due to their ability to glide across agar despite not possessing flagella [1, 2].
C. gingivalis possess all genes needed for glycolysis but are unable to undertake complete oxidative phosphorylation, as seen by the lack of NADH dehydrogenase, cytochrome bc1 and cytochrome c oxidase in its ‘KEGG’ metabolic pathway [9]. However, all genes encoding the bacterial F-type ATP-ase subunits are present, suggesting that the ATP-ase gene product is now non-functional and that it is fermentative [10]. As strictly fermentative chemoorganotrophic facultative anaerobes, C. gingivalis preferentially ferment compounds such as glycogen or starch to produce acidic end products, e.g., acetate and succinate [1].
Metabolism is rapid in aerobic conditions and can still occur in anaerobic conditions provided there are elevated CO2 levels [1].
Ecology
Species of the genus Capnocytophaga have been found in canine dental environments, such as C. canimorsus and C. cynodegmi, however, C. gingivalis is a facultative anaerobe which has been isolated in human supragingival plaque, where it constitutes a part of the normal human oral microbiota [2]. As classified by Socransky and Haffajee in 2002, C. gingivalis are part of the ‘green complex’, which secondarily colonise the human oral cavity and grow on primary colonisers of the Streptococcus genus [11].
Adhesins have been described on the surface of C. gingivalis which allow aggregation of C. gingivalis with other oral bacteria to form a biofilm on the gingiva [12]. Sequenced strains of C. gingivalis have only been isolated from the oral cavity and theoretically, any of capnophilic C. gingivalis’ other potential environments would require high CO2 levels.
Pathology
C. gingivalis is part of the healthy human oral microbiome where it has also been implicated in causing systemic infection.
Recent studies into non-systemic disease caused by C. gingivalis studies have focused on dental caries. In the study by Haffajee et al, 2009, teeth with high plaque mass contained more C. gingivalis , and C. gingivalis was isolated in 14% of over-60s with root caries [13]. Regardless of this correlation between C. gingivalis and plaque/dental caries, recent microbiome analysis by Jiang et al in 2014 suggests no link between C. gingivalis and childhood dental caries [2, 3].
When normal mucosal barriers in the human oropharynx undergoes trauma, disease or ulceration, C. gingivalis can act as an opportunistic pathogen and upregulate genes responsible for the invasive pathogenesis. A significant number of C. gingivalis systemic infections are polymicrobial. Periodontitis, causing chronic inflammation, has also been suggested to promote colonisation by antibiotic resistant resident C. gingivalis bacteria [7]. Capnocytophaga species, once invasive, are able to cause serious systemic infections such as bacteraemia, endocarditis and lung infections in immunocompromised cancer patients [3, 14-16]. One case of C. gingivalis -related bacteraemia was recorded in 1992 in a Hodgkin’s lymphoma patient who had just undergone bone marrow transplantation [17]. This elicited the assemblage of case studies detailing C. gingivalis -related bacteraemia in immunocompromised patients, and led to the increased appreciation for the need for differential diagnosis.
Application to biotechnology
A 2013 study by Ehrmann et al collected 48 subgingival isolates of human oral Capnocytophaga from both healthy and ill patients. In this study, no C. gingivalis was isolated from haematology patients, only from periodontitis patients and healthy volunteers, confirming C. gingivalis makes up a part of the healthy human oral microbiota. Of the 48 Capnocytophaga isolates, 44% had beta lactam resistance genes, and 29% had macrolide-lincosamide-streptogramin (MLS) resistance genes. In contrast, 11% of C. gingivalis isolates had beta lactam resistance genes, and 16% had MLS resistance genes. This study supports the theory that C. gingivalis and related species are a significant reservoir of beta lactams and MLS antibiotic resistance genes. Exploiting the pathway related to antibiotic synthesis could be a future focus for biotechnology, however, other species of Capnocytophaga such as C. ochracea may be more useful in this respect.
Current research
C. gingivalis contains CRISPR elements, and was recently included in the metagenomic data for a 2015 study which characterised CRISPR compositions under different periodontal status [18]. It was found that the composition of direct repeats and spacers were significantly different between patients with chronic periodontitis and those with healthy gingiva. The authors recommended further research into the functionality of CRISPR elements in oral ecology.
Some strains of C. gingivalis have been shown to possess antibiotic resistance to beta lactams and MLS [7]. A case study and review by Ehrmann et al in 2016 investigated the molecular mechanism for fluoroquinolone resistance in C. gingivalis , first described by Geisler et al in 2001 [19]. It was found that DNA gyrase (GyrA) was the primary target for fluoroquinolones in gram negative bacteria. The presumed gyrA quinolone-resistance-determining-region (QRDR) of C. gingivalis was determined, and a Gly80Asn mutation within the QRDR was found in a fluoroquinolone resistant C. gingivalis isolate, supporting the hypothetical QRDR location. This review also highlighted the case of a multidrug resistant strain of C. gingivalis , which acquired resistance to third generation cephalosporins, MLS and fluoroquinolones, and eventually led to systemic infection and COPD manifestation. Further research is recommended to evaluate the frequency of the Gly80Asn QRDR mutation in response to selection pressure.
References
2. Jolivet-Gougeon A. Capnocytophaga species.
5. NCBI. Genome structure of the ATCC 33624 strain of Capnocytophaga gingiva.
9. KEGG. KEGG pathway for glycolysis.
10. KEGG. KEGG pathway for oxidative phosphorylation.
11. Socransky S, AD H. 2002. Dental biofilms: difficult therapeutic targets. Periodontology 28:12-55.
20. Image of colony of C. gingivalis
21. Image of a gram stain of C. gingivalis
- ↑ MICR3004
This page is written by <Simone Webb> for the MICR3004 course, Semester 2, 2016
