Xylella fastidiosa
A Microbial Biorealm page on the genus Xylella fastidiosa
Classification
Higher order taxa
Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Xylella
Species
Xylella fastidiosa
|
NCBI: Taxonomy |
Description and significance
Xylella fastidiosa is a pathogenic bacterium that infects plants, causing a variety of diseases in over 100 plant species, including grapevine, citrus, almonds, coffee, and many other species of economic importance. Among the diseases it causes are Pierce's Disease in grapevine, Citrus Variegated Chlorosis (CVC) in oranges, and leaf scorch diseases in almond, coffee, and oleander. It was first discovered associated with Pierce's Disease in southern California grapevines in 1973, and first grown in culture in 1978 (1). It was then identified as the agent that causes CVC in 1993, a disease first discovered in Brazil in 1987 (2). X. fastidiosa exclusively colonizes the xylem, the water-conducting systems of plants, forming biofilms, and is transmitted from plant to plant by xylem-feeding leafhopper insects, including the glassy-winged sharpshooter. It poses a serious agricultural and economic threat, as it is responsible for major crop losses globally, and is included in the federal government's Agricultural Select Agent list (3). Because of its public importance as an agricultural threat, genomic studies of different host plant strains have been underway in an attempt to gain insight into virulence factors, and consequently the development of microbiological control and disease management strategies.
Genome structure
The genome sequences of four strains of Xylella fastidiosa have been sequenced to date: 9a5c, Ann-1, Dixon, and Temecula-1. Genome comparisons have revealed that the Pierce's Disease strain Temecula-1 genome represents the ancestral genome of Xylella fastidiosa. There are 1,579 homologous genes in all four strains sequenced, accounting for approximately 76.2% of the genome size. All possess pathogenicity-related genes involved in the colonization of their plant host, including a 7kb conserved gene cluster encoding proteins associated with pili biogenesis, functioning in attachment to the host (3), as well as genes for the type II secretion system, which is involved in exporting exoenzymes that degrade plant cell walls, allowing bacteria to colonize. All X. fastidiosa strains also possess an exopolysaccharide similar to the xanthan gum produced by Xanthomonas campestris, and may play a role in biofilm formation. In addition, there are a number of genes unique to each strain: 60 in 9a5c, 83 in Ann-1, 54 in Dixon, and 9 in Temecula-1 (3).
The genome of Xylella fastidiosa 9a5c, first isolated in 1992 from infected twigs derived from Brazilian Valencia sweet oranges and the cause of CVC, consists of a circular chromosome that is 2,679,306 base pairs long. It has 2,766 protein-coding regions. It also contains two circular plasmids: pXF1.3, and pXF51. pXF1.3 is 1,286 base pairs long with 2 protein-coding regions, while pXF51 is 51,158 base pairs long with 64 protein-coding regions (4). Most genes on the pXF51 plasmid are mobile genetic elements or aid in metabolism, but the plasmid does contain 1 virulence-associated protein (5). At least 83 genes are bacteriophage-derived, providing evidence of phage-mediated horizontal gene transfer, and include virulence-associated genes (2).
The genome of Xylella fastidiosa Ann-1, a strain associated with oleander leaf scorch disease, consists of a linear chromosome that is 5,115,342 base pairs long, with 4,660 protein-coding regions. It has no plasmids (4).
The genome of Xylella fastidiosa Dixon, a strain associated with almond leaf scorch disease, consists of a linear chromosome that is 2,622,328 base pairs long, with 2,358 protein-coding regions. It has no plasmids (4).
The genome of Xylella fastidiosa Temecula-1, first isolated in 1998 from an infected California grapevine and the cause of Pierce's Disease of grapevines, consists of a circular chromosome that is 2,519,802 base pairs long. It has 2,034 protein-coding regions. It also contains 1 circular plasmid, pXFPD1.3, which is 1,346 base pairs long and has 2 protein-coding regions (4).
Two strains of X. fastidiosa, M12 and M23, are currently being sequenced for comparative genome analysis.
Cell structure and metabolism
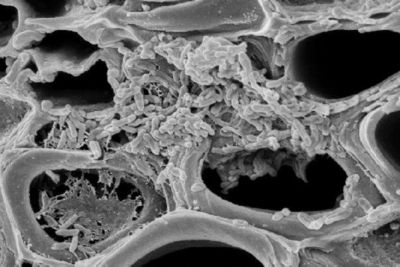
Xylella fastidiosa is a Gram-negative, rod-shaped bacterium with dimensions of 0.25 to 0.35 µm in radius and 0.9 to 3.5 µm in length (6). It has distinctive rippled cell walls, and possesses two types of polar pili: type I pili 0.4 to 1.0 µm in length, and type IV pili 1 to 6 µm in length (6). Both types of pili are positioned at the same pole and aid in xylem attachment, biofilm formation, and twitching motility. X. fastidiosa is a nutritionally fastidious aerobe that grows in the highly specialized environment of the xylem fluid, which contains the lowest concentration of organic energy sources of all plant tissues. However, this nutritionally poor environment does contain specific amino acids, particularly glutamine and asparagine, organic acids, and inorganic ions that are essential nutrient sources for the bacterium, allowing it to efficiently produce energy and grow. Because of its nutrient-poor environment, it has special mechanisms to concentrate and absorb nutrients. It is believed that it possess extracellular polysaccharide glycocayx-like fibers that may function in ion-exchange, nutrient binding within cell aggregations, and conserving and concentrating digestive enzymes released by the bacterium (7).
Ecology
Xylella fastidiosa colonizes the specialized ecological niche of plant xylem vessels. It is able to colonize over 100 species of plants, and its host range continues to expand (8). It can cause a wide variety of scorch diseases in its plant hosts, resulting in widespread crop damage in affected areas, negatively impacting agricultural areas. However, it does not cause disease in most plants. Its residency in the xylem appears to be the main criteria for survival, but it can accumulate and produce disease symptoms if the host becomes weakened or susceptible by drought, root pruning, other diseases, etc. Strains causing Pierce's Disease in California, for example, appear to kill susceptible grapevines in restricted areas called "hot spots", near permanent water sources where leafhopper insect vectors are present (1).
X. fastidiosa forms aggregated biofilms upon colonization of the xylem. Biofilm formation appears to be important for its survival and pathogenicity, and thus there is a connection between aggregation and virulence (1). Colonization of the xylem is also dependent on movement of the bacterium between xylem vessels.
X. fastidiosa can also colonize the foregut of xylem sap-feeding insect vectors such as the sharpshooter. Upon feeding from an infected plant, bacteria enter the insect vector, attaching to the lining of the foregut, where they multiply and form biofilms (7) before being transmitted to another plant host.
Pathology

Xylella fastidiosa causes a variety of plant scorch diseases, most notably Pierce's Disease in grapevines and Citrus Variegated Chlorosis (CVC) in citrus. While there are a number of proposed mechanisms for its pathogenesis, Xylella fastidiosa is thought to cause disease by blocking xylem vessels and water transport, causing water stress and nutritional deficiencies that result in disease symptoms and expression (9). Bacterial pili, exopolysaccharides, degradative enzymes, biofilm formation and cell aggregation, and systematic movement from vessel to vessel all play important factors in pathogen virulence. Polar pili and the secretion of exopolysaccharides mediate plant-bacterium, and bacterium-bacterium adhesion and aggregation, allowing for the formation of cell aggregates which can then cause xylem blockage (2). Its pathogenicity is also dependent upon its ability to move between xylem vessels. This is mediated by twitching motility using polar pili, which allow downward migration in the plant (9), as well as the degradation of the pit membranes (primary cell walls) that protect the xylem by bacterial degradative enzymes. This mechanism of pathogenicity is supported by the fact that avirulent Pierce's Disease strains of X. fastidiosa multiply slowly and are incapable of moving from vessel to vessel (1).
Major symptoms of disease caused by X. fastidiosa include marginal necrosis, leaf abscission, dieback, delayed growth in the spring, and a decline in vigor, which can lead to plant death. Symptoms are usually not visible until either the time of fruit maturation, or in late fall when the host plant is senescing (1). Pierce's Disease-infected grapevines exhibit leaf chlorosis (yellowing) before leaves die progressively, leaving a series of concentric zones of discolored tissue. Fruit clusters may also wilt and dry up (10). CVC can affect all commercial orange trees, causing chlorotic areas on the upper side of leaves with corresponding brown, gummy lesions on the lower side. Affected fruits are small, hardened, and have no commercial value (2).
X. fastidiosa is dependent on its interaction with xylem sap-feeding insect vectors, as well as insect vector interaction with plants, as the insect vectors are necessary for their transmittance to new host plants. They are transferred to the xylem of other host plants by the force of pumping action during feeding of the insect vector, which dislodges some of the bacteria that has colonized in the foregut of the insect, allowing it to enter the xylem (1).
Disease caused by X. fastidiosa is controlled by removing infected shoots by pruning, replanting with healthy plants, the use of insecticides, and the use of resistant cultivars (2).
Application to Biotechnology
There are no compounds or enzymes produced by Xylella fastidiosa currently used for biotechnology. However, the recent characterization of heat shock and thermostable proteins has been of special interest because of their huge potential application to biotechnology, including their use in biochemical assays and cancer research. They are also promising targets for new approaches to combating the diseases the bacterium causes.
Current Research
Because of the huge potential for causing plant and crop destruction, recent research on Xylella fastidiosa has focused on factors that influence its pathogenicity, as well as disease prevention and control mechanisms. In one recent study, genes involved in the heat shock response in X. fastidiosa were investigated by a whole-genome microarray analysis. The heat shock response is induced when cells are exposed to environmentally stressful conditions, such as increased temperature, causing the production of proteins that mediate correct folding of polypeptides, counteracting stress disturbances and allowing the cell to continue to survive and function. Koide et al. (2006) discovered that 261 genes (9.7% of the genome) were induced, and 222 genes (8.3% of the genome) were repressed during the heat shock response in X. fastidiosa. In addition to the induction of classical heat shock genes, an upregulation of virulence-associated genes was observed, including nonfimbrial adhesions and degradative enzymes, which may play an important role in bacterial adhesion in response to stress. It has been observed that CVC disease is more accentuated during warmer months and the disease symptoms are more severe. This suggests that the heat shock response induced upon heat exposure enhances the success of the bacterial infection in the host plant and pathogenesis, contributing to the bacterium's destructive effects (11).
Researchers have also been focusing on bacterial targets for disease control. One study presented the recombinant expression and characterization of a X. fastidiosa cysteine protease called Xylellain in a nonpathogenic strain of the bacterium. It was found that mice immunized with Xylellain produced antibodies that recognized a 31 kDa protein in the 9a5c pathogenic strain of X. fastidiosa. The antibodies did not, however, recognize proteins in the nonpathogenic strain J1a12. This suggests that Xylellain is absent or expressed in low concentrations in nonpathogenic strains, and thus may be an important pathogenic factor for disrupting plant tissue and allowing bacterial spread. This recent discovery has also enabled researchers to identify Xylellain as a possible target for combating CVC and other diseases caused by pathogenic strains of X. fastidiosa (12).
Another recent study has focused on evaluating the effectiveness of antibiotics and antimicrobial peptides against Xylella fastidiosa. Kuzina et al. (2006) tested 12 antibiotics and 18 antimicrobial peptides on 10 strains of X. fastidiosa, and the minimal inhibitory concentration, the lowest dose of the antibiotic or antimicrobial peptide required to inhibit bacterial plate growth, was measured. It was discovered that the antibiotics gentamycin, tetracycline, ampicillin, kanamycin, novobiocin, chloramphenicol, and rifampin had the lowest minimal inhibitory concentration for X. fastidiosa strains, and therefore were the most effective antibiotics against the bacterium. In addition, 4 antimicrobial peptides (Magainin 2, Indolicidin, PGQ, and Dermaseptin) were toxic to all X. fastidiosa strains. This proves that antibiotics and antimicrobial peptides have some activity against the pathogen, and thus have promising applications in the prevention of X. fastidiosa-causing disease in plants (13).
References
4. Entrez Genome Database: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=13864
5. Xylella fastidiosa Genome Project: http://aeg.lbi.ic.unicamp.br/xf/
10. College of Natural Resources, University of California, Berkeley: http://www.cnr.berkeley.edu/xylella/
Edited by Kathryn Thompson, student of Rachel Larsen and Kit Pogliano, University of California, San Diego
