Control of Initiation of Sporulation of Bacillus Subtilis: Difference between revisions
Wendyma321 (talk | contribs) No edit summary |
No edit summary |
||
| (4 intermediate revisions by one other user not shown) | |||
| Line 2: | Line 2: | ||
=Basic Information and Ecological Niche= | =Basic Information and Ecological Niche= | ||
[[Bacillus subtilis]] is a gram-positive, rod-shaped bacterium found in soil and vegetation [http://lib.jiangnan.edu.cn/asm/115-introduce.htm 17] studied for its ability to form [[ | [[Bacillus subtilis]] is a gram-positive, rod-shaped bacterium found in soil and vegetation [http://lib.jiangnan.edu.cn/asm/115-introduce.htm 17] studied for its ability to form [[bacterial endospores]][http://www.britannica.com/EBchecked/topic/47965/bacillus 1]. Because B. subtilis lives in a dynamic soil environment where conditions can switch quickly, it has adapted sporulation to combat these challenges. Alternatively, it may be found in the gastrointestinal tract of animals, where it may commence its virulent life cycle [http://www.ncbi.nlm.nih.gov/pubmed/19068230 10]. | ||
Bacillus subtilis as a Model Organism
B. subtilis is the model organism for sporulation to its widespread availability and easy cultivation. This is due to their ability to tolerate oxygen [http://www.textbookofbacteriology.net/Bacillus.html 19], as opposed to [[Clostridium]], which is anaerobic [http://www.phac-aspc.gc.ca/lab-bio/res/psds-ftss/clostridium-spp-eng.php 5]. Additionally, B subtilis is ubiquitous and broadly adapted to growing in many environments [http://www.ncbi.nlm.nih.gov/pubmed/18467096 7]. | Bacillus subtilis as a Model Organism
B. subtilis is the model organism for sporulation to its widespread availability and easy cultivation. This is due to their ability to tolerate oxygen [http://www.textbookofbacteriology.net/Bacillus.html 19], as opposed to [[Clostridium]], which is anaerobic [http://www.phac-aspc.gc.ca/lab-bio/res/psds-ftss/clostridium-spp-eng.php 5]. Additionally, B subtilis is ubiquitous and broadly adapted to growing in many environments [http://www.ncbi.nlm.nih.gov/pubmed/18467096 7]. | ||
| Line 25: | Line 25: | ||
The first step in the transduction system include [http://en.wikipedia.org/wiki/Histidine_kinase sensor kinases], which autophosphorylate upon receiving a signal. KinA has three methods of regulation: | The first step in the transduction system include [http://en.wikipedia.org/wiki/Histidine_kinase sensor kinases], which autophosphorylate upon receiving a signal. KinA has three methods of regulation: | ||
2. KinA is regulated by Sda, a protein which binds to KinA and barricades the phosphotransfer of KinA to SpoF. Upon DNA damage or blockage in DNA replication, the sda gene is transcribed. Therefore, Sda prevents sporulation via inhibiting phosphotransfer activity of Spo0A when DNA replication is compromised | 1. KinA is controlled by KipI, an inhibitor of kinase activity of KinA, which is counteracted by an anti-inhibitor protein, KipA, and both belong to the same operon. Sufficient amounts of glucose and fixed nitrogen will favour dephosphorylation of KinA via this pathway [http://www.sciencedirect.com/science/article/pii/S0966842X9801350X 12]. | ||
3. KinA has three PAS domains which sense changes in redox potential and oxygen levels. Depending on whether the condition is favourable, the PAS domains influence autophosphorylation | |||
2. KinA is regulated by Sda, a protein which binds to KinA and barricades the phosphotransfer of KinA to SpoF. Upon DNA damage or blockage in DNA replication, the sda gene is transcribed. Therefore, Sda prevents sporulation via inhibiting phosphotransfer activity of Spo0A when DNA replication is compromised [http://www.ncbi.nlm.nih.gov/pubmed/15556029 15]. | |||
3. KinA has three PAS domains which sense changes in redox potential and oxygen levels. Depending on whether the condition is favourable, the PAS domains influence autophosphorylation [http://www.ncbi.nlm.nih.gov/pubmed/15556029 15]. | |||
=Regulation of Spo0F= | =Regulation of Spo0F= | ||
The regulation of response regulatory protein Spo0F has three levels of control - Rap phosphatases are controlled by the PhrA proteins, which are controlled by CodY proteins. | The regulation of response regulatory protein Spo0F has three levels of control - Rap phosphatases are controlled by the PhrA proteins, which are controlled by CodY proteins. | ||
RapA, B and E are phosphatases which dephosphorylate Spo0F by binding onto it. RapA, B and E are regulated by pentapeptides PhrA, PhrB, and PhrE proteins. Phr peptides sense cell density and cause inhibition of Rap proteins which cause dephosphorylation of Spo0F~P and promoting sporulation | RapA, B and E are phosphatases which dephosphorylate Spo0F by binding onto it. RapA, B and E are regulated by pentapeptides PhrA, PhrB, and PhrE proteins. Phr peptides sense cell density and cause inhibition of Rap proteins which cause dephosphorylation of Spo0F~P and promoting sporulation [http://www.ncbi.nlm.nih.gov/pubmed/15556029 15]. Under low cell density conditions, it may be more beneficial to wait for food than sporulating. The Phr proteins are first transported outside the cell then imported back in again after cleavage, upon which it will inhibit the Rap proteins’ phosphatase activity [http://www.sciencedirect.com/science/article/pii/S0966842X9801350X 12]. This timing mechanism is used to ensure that B. Subtilis has had an opportunity to explore other possibilities before committing to sporulation [http://www.ncbi.nlm.nih.gov/pubmed/10354595 11]. | ||
Furthermore, Cod Y, a sensor of guanine nucleotide levels, inhibits PhrA which inhibits RapB which dephosphorylates Spo0F. A sharp decrease in GTP and GDP levels causes relief of | Furthermore, Cod Y, a sensor of guanine nucleotide levels, inhibits PhrA which inhibits RapB which dephosphorylates Spo0F. A sharp decrease in GTP and GDP levels causes relief of | ||
CodY-mediated repression of phrA genes, causing sporulation in otherwise non-sporulating conditions | CodY-mediated repression of phrA genes, causing sporulation in otherwise non-sporulating conditions [http://www.ncbi.nlm.nih.gov/pubmed/15556029 15]. | ||
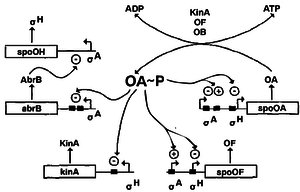
=Regulation of Spo0A= | =Regulation of Spo0A= | ||
The regulation of Spo0A is controlled via three pathways – AbrB, autoregulation and Sin proteins. | The regulation of Spo0A is controlled via three pathways – AbrB, autoregulation and Sin proteins. | ||
Firstly, when Spo0A is phosphorylated, it represses the abrB promoter. The AbrB protein, when present, represses the σH promoter which forms part of the RNA polymerase needed to transcribe sporulation genes | Firstly, when Spo0A is phosphorylated, it represses the abrB promoter. The AbrB protein, when present, represses the σH promoter which forms part of the RNA polymerase needed to transcribe sporulation genes [http://www.ncbi.nlm.nih.gov/pubmed/8257105 9]. Therefore phosphorylation of SpoA activates early sporulation genes. | ||
Secondly, tying into this pathway is the autoregulation of Spo0A. In vegetative state, a vegetative σ factor σA | Secondly, tying into this pathway is the autoregulation of Spo0A. In vegetative state, a vegetative σ factor σA [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC372899/ 3] transcribes spo0A and spo0F genes at a very low level even in the excess of nutrients. At the start of transition state (between vegetative and sporulation), there is a rapid rise in the production of Spo0F and Spo0A as a result of a positive feedback loop where the existing Spo0F~P and Spo-0A~P positively regulate spo0A and spo0F genes. This allows for rapid initiation of sporulation in cases of sudden shift from nutrient-excess to nutrient-poor conditions. When sufficient amounts of Spo0A~P and Spo0F~P is achieved, the vegetative promoter of both genes is inhibited and the downstream σH promoter is activated, which causes early sporulation genes to be transcribed [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC98955/ 6]. | ||
Thirdly, SinR is a protein that binds to the promoter site of spo0A gene and represses transcription. Suppression is relieved by SinI, a protein that is induced as cells enter the stationary phase | Thirdly, SinR is a protein that binds to the promoter site of spo0A gene and represses transcription. Suppression is relieved by SinI, a protein that is induced as cells enter the stationary phase [http://www.sds.vanderbilt.edu/singleton/BSCI275B/sporulationinitiation.pdf 16]. | ||
[[File:Spo0A.png|300px|thumb|right|Regulatory Pathways of Spo0A | [[File:Spo0A.png|300px|thumb|right|Regulatory Pathways of Spo0A [http://www.ncbi.nlm.nih.gov/pubmed/8257105 9]]] | ||
=Initiation of Sporulation= | =Initiation of Sporulation= | ||
The purpose of these signals is to phosphorylate Spo0A which would activate various genes in order to initiate sporulation. In sporulation, unequal cell division results in an endospore encased in a large mother cell. The mother cell eventually dies and lyses, and the endospore becomes an environmental-resistant spore. Once the conditions become favourable again, the endospore will resume vegetative cycle | The purpose of these signals is to phosphorylate Spo0A which would activate various genes in order to initiate sporulation. In sporulation, unequal cell division results in an endospore encased in a large mother cell. The mother cell eventually dies and lyses, and the endospore becomes an environmental-resistant spore. Once the conditions become favourable again, the endospore will resume vegetative cycle [http://www.devbio.biology.gatech.edu/?page_id=15 4]. | ||
=References= | =References= | ||
Latest revision as of 18:45, 29 September 2015
Basic Information and Ecological Niche
Bacillus subtilis is a gram-positive, rod-shaped bacterium found in soil and vegetation 17 studied for its ability to form bacterial endospores1. Because B. subtilis lives in a dynamic soil environment where conditions can switch quickly, it has adapted sporulation to combat these challenges. Alternatively, it may be found in the gastrointestinal tract of animals, where it may commence its virulent life cycle 10. Bacillus subtilis as a Model Organism B. subtilis is the model organism for sporulation to its widespread availability and easy cultivation. This is due to their ability to tolerate oxygen 19, as opposed to Clostridium, which is anaerobic 5. Additionally, B subtilis is ubiquitous and broadly adapted to growing in many environments 7.
Introduction into Endospores
One of the features of B. subtilis is its ability to sporulate, or form endospores which are a dormant form of a bacterium which is metabolically inactive 1, formed in conditions of stress and can persist in a hostile conditions (ie lack of nutrients, etc.) for long periods of time until conditions become favourable again.
Applications of Endospore Research
The use of B. subtilis to study sporulation has many applications, as research may shed light on various other spore formers which have been used as natural insecticides, exploited in production of enzymes, antibiotics and probiotics, served as vectors for drug delivery and vaccines 2, and have been the subject of biological warfare 13.
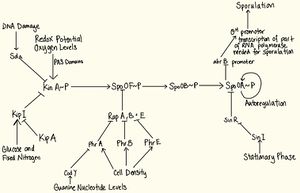
The phosphorelay system
The phosphorelay system consists of four basic reactions. The first event is a signal from the environment which causes KinA, a sensor kinase (or other kinases) to autophosphorylate 9. The phosphate group is transferred to Spo0F, which is the response regulator 3. The transfer of phosphate from Spo0F~P to Spo0A is mediated via a phosphorylated enzyme intermediate Spo0B. The phosphorylated Spo0A goes on to activate the genes necessary for sporulation. The networks of signals which regulate the phosphotransfer network receives input from many sources so that the proper choice is made using an integrated view of the environmental and metabolic conditions of the bacterium 18. The signals covered in this paper include availability of glucose and fixed nitrogen, DNA damage, oxygen levels, redox potential, and stationary phase. variability.
Regulation of Sensor Kinases
The first step in the transduction system include sensor kinases, which autophosphorylate upon receiving a signal. KinA has three methods of regulation:
1. KinA is controlled by KipI, an inhibitor of kinase activity of KinA, which is counteracted by an anti-inhibitor protein, KipA, and both belong to the same operon. Sufficient amounts of glucose and fixed nitrogen will favour dephosphorylation of KinA via this pathway 12.
2. KinA is regulated by Sda, a protein which binds to KinA and barricades the phosphotransfer of KinA to SpoF. Upon DNA damage or blockage in DNA replication, the sda gene is transcribed. Therefore, Sda prevents sporulation via inhibiting phosphotransfer activity of Spo0A when DNA replication is compromised 15.
3. KinA has three PAS domains which sense changes in redox potential and oxygen levels. Depending on whether the condition is favourable, the PAS domains influence autophosphorylation 15.
Regulation of Spo0F
The regulation of response regulatory protein Spo0F has three levels of control - Rap phosphatases are controlled by the PhrA proteins, which are controlled by CodY proteins. RapA, B and E are phosphatases which dephosphorylate Spo0F by binding onto it. RapA, B and E are regulated by pentapeptides PhrA, PhrB, and PhrE proteins. Phr peptides sense cell density and cause inhibition of Rap proteins which cause dephosphorylation of Spo0F~P and promoting sporulation 15. Under low cell density conditions, it may be more beneficial to wait for food than sporulating. The Phr proteins are first transported outside the cell then imported back in again after cleavage, upon which it will inhibit the Rap proteins’ phosphatase activity 12. This timing mechanism is used to ensure that B. Subtilis has had an opportunity to explore other possibilities before committing to sporulation 11. Furthermore, Cod Y, a sensor of guanine nucleotide levels, inhibits PhrA which inhibits RapB which dephosphorylates Spo0F. A sharp decrease in GTP and GDP levels causes relief of CodY-mediated repression of phrA genes, causing sporulation in otherwise non-sporulating conditions 15.
Regulation of Spo0A
The regulation of Spo0A is controlled via three pathways – AbrB, autoregulation and Sin proteins. Firstly, when Spo0A is phosphorylated, it represses the abrB promoter. The AbrB protein, when present, represses the σH promoter which forms part of the RNA polymerase needed to transcribe sporulation genes 9. Therefore phosphorylation of SpoA activates early sporulation genes. Secondly, tying into this pathway is the autoregulation of Spo0A. In vegetative state, a vegetative σ factor σA 3 transcribes spo0A and spo0F genes at a very low level even in the excess of nutrients. At the start of transition state (between vegetative and sporulation), there is a rapid rise in the production of Spo0F and Spo0A as a result of a positive feedback loop where the existing Spo0F~P and Spo-0A~P positively regulate spo0A and spo0F genes. This allows for rapid initiation of sporulation in cases of sudden shift from nutrient-excess to nutrient-poor conditions. When sufficient amounts of Spo0A~P and Spo0F~P is achieved, the vegetative promoter of both genes is inhibited and the downstream σH promoter is activated, which causes early sporulation genes to be transcribed 6. Thirdly, SinR is a protein that binds to the promoter site of spo0A gene and represses transcription. Suppression is relieved by SinI, a protein that is induced as cells enter the stationary phase 16.
Initiation of Sporulation
The purpose of these signals is to phosphorylate Spo0A which would activate various genes in order to initiate sporulation. In sporulation, unequal cell division results in an endospore encased in a large mother cell. The mother cell eventually dies and lyses, and the endospore becomes an environmental-resistant spore. Once the conditions become favourable again, the endospore will resume vegetative cycle 4.
References
[1] Bacillus. (2013). Retrieved October 31, 2013 from http://www.britannica.com/topic/47965/bibliography
[2] Barak, I. (2005). From fundamental studies of sporulation to applied spore research. Molecular Microbiology, 55(2): 330-338.
[3] Burbulys, D. (1991) Initiation of Sporulation in B. subtilis is Controlled by a Multicomponent Phosphorelay. Cell Press, 64: 545-552.
[4] Choi, J. (2009) Bacillis subtilis sporulation. Developmental Biology Interactive. http://www.devbio.biology.gatech.edu/?page_id=15. Accessed 19 November 2013.
[5] Clostridium Spp. Pathogen Safety Data Sheet – Infectious Substances. (2012) Retrieved November 18, 2013, from Public Health Agency of Canada, website: http://www.phac- aspc.gc.ca/lab-bio/res/psds-ftss/clostridium-spp-eng.php
[6] Driks, A. (1999). Bacillus subtilis Spore Coat. Microbiology and Molecular Biology Reviews, 63 (1): 1-20.
[7] Earl, A. (2008). Ecology and genomics of Bacillus subtilis. Trends in Microbiology, 16 (6): 269- 275.
[8] Errington, J. (1993). Bacillus subtilis Sporulation: Regulation of Gene Expression and Control of Morphogenesis. Microbiological Reviews, 57 (1): 1-33.
[9] Hoch, J. (1993) Regulation of the Phosphorelay and the initiation of sporulation in Bacillus subtillis. Annual Review of Microbiology 47: 441-465.
[10] Hong, H. (2009) Bacillus subtilis isolated from the human gastrointestinal tract. Research in Microbiology, 160 (2): 134-43.
[11] Msadek, M. (1999) When the going gets tough: survival stretegies and environment signaling networks in Bacillus subtilis. Trends in Microbiology, 7(5): 201-207.
[12] Perego, M. (1998) Kinases-phosphatase competition regulates Bacillus subtilis development. Trends in Microbiology, 6 (9): 366-370.
[13] Pezim, M. (2013) Anthrax as a biological weapon. MediResource Inc. http://chealth.canoe.ca/channel_section_details.asp?text_id=1484&channel_id=1020&relation_id=71020. Accessed 18 November 2013.
[14] Piggot, P. (1996) Spore Development in Bacillus subtilis. Current Opinion in Genetics & Development, 6: 531-537.
[15] Piggot, P. (2004) Sporulation of Bacillus subtilis. Current Opinion in Microbiology 2004, 7:579-586.
[16] Sonenshein, A. (2000). Control of sporulation initiation in Bacillus subtilis. Current Opinion in Microbiology 2000, 3: 561-566.
[17] Smith, A. (2011). Gram Stain: Gram-Positive Rods. Atlas. American Society for Microbiology, Washington, DC. www.microbelibrary.org Accessed 31 October 2013.
[18] Strauch, M. (1993). Signal transduction in Bacillus subtilis sporulation. Current Opinion in Genetics and Development, 3: 203-212.
[19] Todar, K. (2012). The Genus Bacillus. Todar’s Online Textbook of Bacteriology.www.textbookofbacteriology.net/Bacillus.html. Accessed 18 November 2013.