Anellovirus: Difference between revisions
| Line 80: | Line 80: | ||
[https://jidc.org/index.php/journal/article/view/26142664/1314 Manzin A, Mallus F, Macera L, Maggi F, Blois S (2015) Global impact of Torque teno virus infection in wild and domesticated animals. Journal of Infection in Developing Countries 9(6): 562-570] | [https://jidc.org/index.php/journal/article/view/26142664/1314 Manzin A, Mallus F, Macera L, Maggi F, Blois S (2015) Global impact of Torque teno virus infection in wild and domesticated animals. Journal of Infection in Developing Countries 9(6): 562-570] | ||
[https://www.nature.com/articles/bmt2015262 Masouridi-Levrat S, Pradier A, Simonetta F, Kaiser L, Chalandon Y, Roosnek E (2016) Torque teno virus in patients undergoing allogeneic hematopoietic stem cell transplantation for hematological malignancies. Bone Marrow Transplantation 51(3): 440-442] | |||
[https://www.ncbi.nlm.nih.gov/pubmed/9405239/ Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M (1997) A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochemical and Biophysical Research Communications 241(1): 92-97] | [https://www.ncbi.nlm.nih.gov/pubmed/9405239/ Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M (1997) A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochemical and Biophysical Research Communications 241(1): 92-97] | ||
Revision as of 14:33, 23 April 2018
Introduction
By Julia Josowitz
At right is a sample image insertion. It works for any image uploaded anywhere to MicrobeWiki. The insertion code consists of:
Double brackets: [[
Filename: Anellotree.jpg
Thumbnail status: |thumb|
Pixel size: |300px|
Placement on page: |right|
Legend/credit: Electron micrograph of the Ebola Zaire virus. This was the first photo ever taken of the virus, on 10/13/1976. By Dr. F.A. Murphy, now at U.C. Davis, then at the CDC.
Closed double brackets: ]]
Other examples:
Bold
Italic
Subscript: H2O
Superscript: Fe3+
Overview
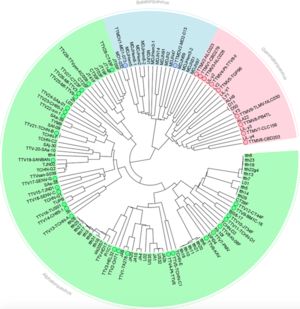
Anelloviruses are small, circular, single stranded, negative-sense DNA viruses found in blood plasma. While they are not known to cause any harm, high viral loads are associated with immune suppression and diseases such as hepatitis, cancer, and autoimmune diseases (Blatter et al.).The first anellovirus discovered was torque teno virus (TTV) in 1997, which was initially believed to be part of the pathology of hepatitis, but since then it has been found that these viruses are relatively widespread and heterogenous, with no known pathogenenicity (Spandole et al.). Within the family Anelloviridae there are three known genera: Alphatorquevirus, Betatorquevirus, and Gammatorquevirus. Alphatorquevirus includes TTV, while Betatorquevirus includes torque teno mini virus and Gammatorquevirus includes torque teno midi virus. Anelloviruses that infect animals have also been found in cats, dogs, and even pigs. TTVs are very prevalent in the human population, with more than 80% of the population infected, and appear to establish persistent infections, which raises the question of how they evade the host immune system, and almost suggest a commensualist relationship.
Anelloviruses are unenveloped viruses that have a capsid made of 12 pentameric capsomers. Virion particle sizes range from 30 to 50 nm for TTV, but are less than 30 nm for torque teno mini virus (TTMV). The crystal structure of TTV has not been solved and these viruses cannot be grown in vitro due to lack of compatible cell systems. Double stranded replicative intermediates have been found in the bone marrow and liver of humans, suggesting that these cells support replication of the virus. Recent studies have also suggested that the virus may be able to replicate in peripheral blood mononuclear cells under certain conditions (Thom and Petrik).
While the main transmission route is believed to be blood, there is evidence of multiple other routes of transmission. Because anelloviruses are present in pharyngeal mucus, it has been suggested that they are also disseminated via the respiratory tract and salivary droplets. Anelloviruses are present in the fecal matter of infected organisms and have been detected in sewage water, as well as in some meat products, which suggests an oral route of transmission. Anelloviruses have been found in the cervical epithelium, where they replicate to high numbers, suggesting a route of sexual transmission. It is possible that viruses are transmitted from mother to offspring, as the viruses have been found in amniotic fluid and umbilical cord blood, and the newborn viral genotype usually resembles the mother. This could also help explain the prevalence of the viruses in populations. However, it is also possible that newborns become infected early in life simply through contact with the mother. (Spandole)
Genome
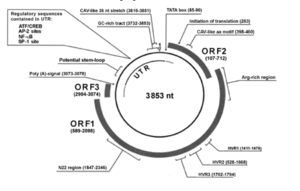
The anellovirus genome is made of a circular single stranded DNA molecule that has a GC rich region of 117 nucleotides. Genome size varies, but the TTV genome ranges from 3.6 to 3.9 kb. Of this, there is a .2 kb untranslated region and a 2.6 kb coding region. The untranslated region has been highly conserved, suggesting that it likely plays a role in viral replication. Interestingly, the coding region has two large open reading frames, the first of which encodes 770 amino acid residues and another that encodes large peptides in TTV. Anellovirus is hypothesized to replicate using the rolling-circle method. The genome of anellovirus is very heterogenous, with high divergence. Different genotypes can infect the same host at the same time, even within the same tissue. Researchers hypothesize that this may be due to a high mutation rate in TTV, which is uncommon for DNA viruses, which use the host's own replicative machinery, which is much less error prone. However, research in single stranded DNA viruses such as parvoviruses has found that the single stranded nature of the genome and its ability to encode replication proteins may account for high rates of mutation. Thus, this could explain the high rates of mutation in Anelloviruses.
Recently, it has also been proposed that intra-genomic recombination could account for the heterogeneity of the Anellovirus genome. It has been suggested that TTV has co-evolved with the host for over millions of years, allowing for great variability.
Potential interaction with cytokine signaling
Other DNA viruses are known to stimulate immune cells to produce inflammatory cytokines, so Rocchi et al. studied whether TTVs could also have a direct effect on immune system signaling. They used lipofectin, which incorporates DNA and protects it from nucleases, to simulate a viral capsid. Spleen cells were treated with lipofectin coupled with TTV DNA, which resulted in an increase in inflammatory cytokines. These results suggest that TTV DNA can directly activate transcription of inflammatory genes within target cells. Inhibitors were used to determine through which receptor TTV DNA was able to change host transcription. Toll-like receptor 9 was found to be the main mediator of the TTV DNA interaction.
Anelloviruses have also been suggested to modulate the host immune system using micro RNAs (miRNAs). miRNAs are small noncoding RNAs (around 22 nucleotides) that play a role in posttranscriptional gene regulation. Viruses from the herpesvirus and retrovirus families have been found the encode viral miRNAs, and these viruses are also characterized by the ability to establish a persistent infection in their host, much like anelloviruses. Using computer simulations and synthetic techniques, Kincaid et al. was able to demonstrate anelloviruses likely encode viral miRNAs. It has also been suggested that viral miRNAs may play a role in immune evasion.
Kincaid et al. developed computational methods to predict the possibility of miRNAs from the primary sequence of genes. They identified a diverse group of miRNAs encoded by TTVs, and by engineering recombinant TTVs capable of expressing an RNA transgene in addition to the miRNA, they were able to identify a direct target of TTV miRNA, the gene N-myc interactor (NMI). NMI is a gene that modulates interferon and cytokine signaling. It is stimulated by interferon and has previously been associated with viral induced apoptosis. The authors suggest that the TTV miRNA helps the virus modulate the immune system, by targeting NMI to inhibit interferon and promote immune evasion. However, while strong possible miRNA candidates were observed in all five groups of human TTVs, none were observed in TTMDVs or TTMVs.
Correlation with transplant outcomes
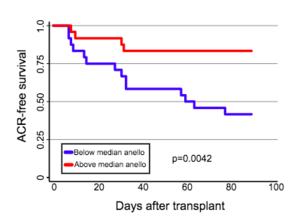
Recent research suggests that anellovirus loads could be used as a predictor of transplant outcomes. High levels of anellovirus are associated with immune suppression, while low levels are correlated with immunocompetence. It had previously been found that lung transplant patients had increased anellovirus levels in their bronchoalveolar fluid and in their serum (Young et al.). Additionally, low plasma anellovirus loads have been found to be associated with acute rejection in heart transplant recipients. This makes anelloviruses a potential marker of chronic rejection, as low plasma anellovirus levels may suggest later rejection.
Blatter et al. investigated the impact of both alphatorqueviruses and betatorqueviruses on the outcome of lung transplants in a pediatric cohort of 57 children. They hypothesized that high levels of anellovirus in the blood would be associated with lower levels of rejection and an overall better outcome. Additionally, they predicted that different genera of anellovirus might be associated with different outcomes, as previous research found that alphatorqueviruses correlated with fever in children but betatorqueviruses did not.
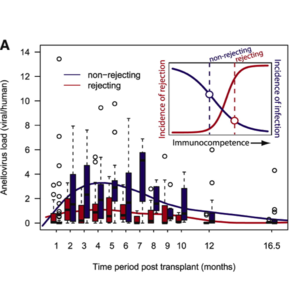
Blatter et al. found that alphatorquevirus levels were higher than betatorquevirus levels in all of the patients, but the load of each increased over the course of the study, which lasted 18 months post transplant. They found that low alphatorquevirus load were associated with acute rejection, while low betatorquevirus levels were correlated with negative long term outcomes, such as death or retransplant. Similar results were found previously by De Vlaminck et al., who observed that from 1 to 16.5 months post-transplant, patients with rejecting outcomes consistently had a lower anellovirus load than non-rejecting patients. Blatter et al. also found distinct phylogenetic clusters of anelloviruses in patients pre-transplant and in controls when compared to post-transplant patients. It is still unclear whether anelloviruses directly interact with the immune system, but this study suggests that anellovirus load can still be a useful biomarker for prediction of transplant outcomes. Additionally, as discussed above, there is evidence that anelloviruses may be able to indirectly effect the immune system via miRNAs.
Interestingly, it appears that while anellovirus loads increase post-transplant and higher loads are correlated with better transplant outcomes, in general anellovirus load correlates with immunosuppression. As a result, it has also been found to correlate with the progression of AIDS in HIV-infected patients. Thom and Petrik found that patients with AIDS consistently had higher anellovirus loads than HIV positive and negative patients. They also found an inverse relationship between the levels of CD4 T cells and anellovirus loads, suggesting that higher viral loads promote immunosuppression.
In patients receiving high dose chemotherapy coupled with autologous stem cell transplantation, an increase in anellovirus load is observed post treatment. Additionally, this increase correlated with an increase in CD8+ T cells, which can help fight cancer. Anelloviral loads returned to baseline around 100 days post transplant, and the authors used this to hypothesize about when patients have returned to immunocompetent status. Anellovirus loads offer an opportunity for clinicans to make informed decisions about when their patients will have the best outcomes for transplants (Focosi et al.).
Conclusion
Overall paper length should be 3,000 words, with at least 3 figures.
References
Edited by Julia Josowitz of Joan Slonczewski for BIOL 238 Microbiology, 2018, Kenyon College.