Faecalibacterium prausnitzii: Difference between revisions
| (43 intermediate revisions by the same user not shown) | |||
| Line 10: | Line 10: | ||
Family: ''Ruminococcaceae'' | Family: ''Ruminococcaceae'' | ||
[ | [[http://www.ncbi.nlm.nih.gov/Taxonomy/ NCBI] link to find] | ||
===Species=== | ===Species=== | ||
| Line 22: | Line 22: | ||
==Description and Significance== | ==Description and Significance== | ||
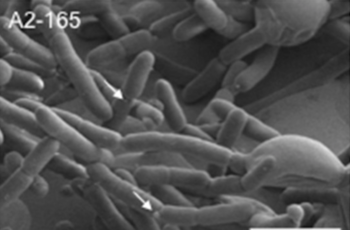
''Faecalibacterium prausnitzii'' | [[File:FPphoto.png|350px|thumb|left|Figure 1. ''Faecalibacterium prausnitzii'' A2-165 strain [18]]] | ||
''Faecalibacterium prausnitzii'' is a rod-shaped, non-motile, non-spore forming, strictly anaerobic bacterium [4, 7]. Its optimal growth temperature is approximately 37 °C [4], which parallels the average temperature of mammals. ''F. prausnitzii'' is extremely oxygen sensitive. Studies have shown that when exposed to ambient air, ''F. prausnitzii'' loses its viability within approximately 2 minutes [11, 6]. These attributes have led to challenges regarding the cultivation and preservation of ''F. prausnitzii'' [12]. | |||
''Faecalibacterium prausnitzii'' colonization in the colon is considered a notable biomarker for a healthy human gastrointestinal tract, and plays a key role in the maintenance of overall gut homeostasis. In | ''Faecalibacterium prausnitzii'' colonization in the colon is considered a notable biomarker for a healthy human gastrointestinal tract, and plays a key role in the maintenance of overall gut homeostasis. In the healthy human gut, ''F. prausnitzii'' comprises around 5% of the gut microbiota [7, 10, 16, 25], and has been detected at levels up to 15% [7]. Decreased abundance of ''F. prausnitzii'' has been linked to gut inflammation and several bowel diseases, including Crohn’s disease, ulcerative colitis, and colorectal cancer [7, 16, 24, 25]. | ||
Because of its significant role in gut homeostasis, ''F. prausnitzii'' strains are of high interest as a next-generation probiotic. Recent research has focused on molecules and strategies aimed at maintaining ''F. prausnitzii'' viable | Because of its significant role in gut homeostasis, ''F. prausnitzii'' strains are of high interest as a next-generation probiotic [18, 26]. Recent research has focused on molecules and strategies aimed at maintaining ''F. prausnitzii'' viable when exposed to varying levels of oxygen. There have been encouraging studies showing ''F. prausnitzii'' maintaining its viability for 24 hours under ambient air, by exploiting extracellular antioxidants such as riboflavin and cysteine, among others [12, 10]. These studies are part of an effort to make ''F. prausnitzii'' commercially viable as a biotherapeutic remedy and probiotic to help those suffering from bowel diseases [13, 16, 19]. Other researchers have also seen the potential of ''F. prausnitzii'' in the agricultural industry as a probiotic for cattle [8, 9]. | ||
==Genome Structure== | ==Genome Structure== | ||
''Faecalibacterium prausnitzii'' has two main phylogroups: Phylogroup I and Phylogroup II. Within these phylogroups, there are different strains whose genomes have been documented in public databases with varying levels of assembly and annotation quality. The genome size of''Faecalibacterium prausnitzii'' exhibits variation, ranging from 2.68 million base pairs (Mbp) to 3.42 Mbp and having wide range of G-C content varying from 54.9% to 63.0%. Most ''F. prausnitzii'' genomes have been constructed from draft assemblies. However one of the first strains to have a complete genome representation is ''F. prausnitzii'' strain A2-165 (Phylogroup II). This strain of ''F. prausnitzii'' has a circular genome containing 3.11 Mbp, 56.3% G-C content, 3,017 total genes, 2,790 coding genes, and 85 RNA genes. | ''Faecalibacterium prausnitzii'' has two main phylogroups: Phylogroup I and Phylogroup II [16]. Within these phylogroups, there are different strains whose genomes have been documented in public databases "with varying levels of assembly and annotation quality" [7]. The genome size of ''Faecalibacterium prausnitzii'' exhibits variation, ranging from 2.68 million base pairs (Mbp) to 3.42 Mbp and having wide range of G-C content, varying from 54.9% to 63.0% [7]. Most ''F. prausnitzii'' genomes have been constructed from draft assemblies. However, one of the first strains to have a complete genome representation is ''F. prausnitzii'' strain A2-165 (Phylogroup II) [7]. This strain of ''F. prausnitzii'' has a circular genome containing 3.11 Mbp, 56.3% G-C content, 3,017 total genes, 2,790 coding genes, and 85 RNA genes [3]. | ||
In a study by Fitzgerald et al. (2018), 31 genomes of high-quality draft as well as complete genomes were used in a comparative genomics analysis to observe intraspecies diversity. The results revealed a high level of genome plasticity and a relatively low level of average nucleotide identity (ANI) between ''F. prausnitzii'' groups. Based on this and other observations, Fitzgerald et al. have proposed to separate ''Faecalibacterium prausnitzii'' into two new species level taxa. | In a study by Fitzgerald et al. (2018), 31 genomes of high-quality draft as well as complete genomes were used in a comparative genomics analysis to observe intraspecies diversity. The results revealed a high level of genome plasticity and a relatively low level of average nucleotide identity (ANI) between ''F. prausnitzii'' groups. Based on this and other observations, Fitzgerald et al. have proposed to separate ''Faecalibacterium prausnitzii'' into two new species-level taxa [7]. | ||
==Cell Structure, Metabolism and Life Cycle== | ==Cell Structure, Metabolism and Life Cycle== | ||
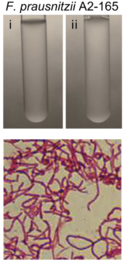
''Faecalibacterium prausnitzii'' shows irregular staining, but typically stains like a Gram-negative bacteria. However, ''F. prausnitzii'' exhibits dermis characteristics that resemble Gram-positive bacteria, | [[File:A2165GS.png|125px|thumb|right|Figure 2. ''F. prausnitzii'' A2-165 Gram-stain [29]]] | ||
''Faecalibacterium prausnitzii'' shows irregular staining, but typically stains like a Gram-negative bacteria [4]. However, ''F. prausnitzii'' exhibits dermis characteristics that resemble Gram-positive bacteria, such as those in clostridial cluster IV (''Clostridium leptum'' group) [6, 17]. Typical Gram-negative bacteria have LPS (lipopolysaccharide) on their outer membranes. LPS proteins are ligands for the transmembrane protein, TLR4 (Toll-like receptor 4). These are not found on ''F. prausnitzii'' or other bacteria in clostridial cluster IV. This suggests that ''F. prausnitzii'' is one of the unique bacteria whose dermis structure does not necessarily align with its Gram-stain [17]. | |||
''F. prausnitzii'' is an acetate-consuming and butyrate-producing bacterium [27, 16]. Although it is one of the most common bacteria found in the human colon, it has been revealed that ''F. prausnitzii'' needs the presence of ''B. thetaiotamicron'', a primary acetate-producer, in order to colonize the colon [27]. ''F. prausnitzii'' produces the short-chain fatty acid (SCFA) butyrate as a product of fermenting indigestible fiber [11]. Butyrate serves as one of the primary energy sources for colonocytes (cells lining the colon) [10] and is a principal molecule that works through various signaling pathways to prevent inflammation, regulate cell proliferation (cancer chemopreventative activities) [22], and maintain the integrity of the colon epithelial mucosa [16, 18, 19]. | |||
==Ecology and Pathogenesis== | ==Ecology and Pathogenesis== | ||
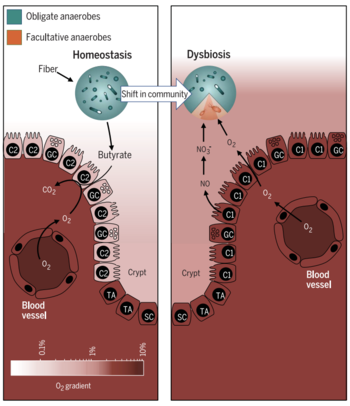
''Faecalibacterium prausnitzii'' is found ubiquitously in the gastrointestinal tracts of both animal and human hosts, and acts as a symbiotic, commensal bacterium within its host. One of the ways in which ''F. prausnitzii'' | [[File:dysbiosis.png|350px|thumb|left|Figure 3. Dysbiosis in the colon [14]]] | ||
''Faecalibacterium prausnitzii'' is found ubiquitously in the gastrointestinal tracts of both animal and human hosts, and acts as a symbiotic, commensal bacterium within its host [16, 19]. One of the ways in which ''F. prausnitzii'' helps maintain a healthy colon is by the production of butyrate via the fermentation of fiber [10, 11]. Butyrate has demonstrated to have various attributes that positively contribute to colonic wellbeing and maintenance [14, 28]. | |||
Butyrate acts as a primary metabolite that maintains a symbiotic relationship between ''F. prausnitzii'' and colonocytes. As ''F. prausnitzii'' produces butyrate, | Butyrate acts as a primary metabolite that maintains a symbiotic relationship between ''F. prausnitzii'' and colonocytes [10]. As ''F. prausnitzii'' produces butyrate, colonocytes respond with increased metabolic activity and cell-sustaining maintenance. These cellular activities deplete oxygen from the lumen of the colon, creating a hypoxic environment for obligate anaerobes to dominate [14]. As a result, the fermentation of fiber into short-chain fatty acids, which are a primary source of colonocyte energy, continues in a symbiotic manner [10, 16]. Higher levels of oxygen in the colon deplete the integrity of the microbiome and result in increased abundance of facultative anaerobic bacteria [14]. This is a tell-tale sign of a gut in dysbiosis and leads to greater inflammation and chronic irritable bowel diseases [14]. Current research has also focused on identifying different ''Faecalibacterium prausnitzii'' strains in patients as a biomarker, in order to discriminate between different gut diseases, hoping to lead to improved diagnostics and treatment [16]. | ||
'' | ''Faecalibacterium prausnitzii''’s further promotes gut health by the production and promotion of anti-inflammatory molecules. These molecules include a proteinaceous “microbial anti-inflammatory molecule” (MAM) that can block the NF-κB pathway of inflammatory response [16]. Other anti-inflammatory attributes observed from ''F. prausnitzii'' include the ability to upregulate anti-inflammatory cytokines, such as IL-10, downregulatre pro-inflammatory molecules such as IL-12 and IFN-γ, and “reduce the severity of acute, chronic, and low-grade chemical-induced inflammation" [16, 22, 25]. ''F. prausnitzii'' has been shown to attenuate the severity of inflammation by inducing the production of tight-junction proteins found in the intestinal mucosa. This anti-inflammatory activity maintains the integrity of the gut lining and prevents leaky gut and further inflammation [16]. Finally, ''F. prausnitzii'' has been found to play a role in regulating proper proportionality of cell types in the gut epithelial lining, leading to proper cell maintenance functions (such as apoptosis) for the evasion of inflammation and cancerous cells [15]. | ||
Rates of depression and anxiety have been associated with inflammatory bowel diseases such as Crohn’s disease and ulcerative colitis. This may be related to depletion of ''F. prausnitzii'' in the microbiome, | Rates of depression and anxiety have been associated with inflammatory bowel diseases such as Crohn’s disease and ulcerative colitis [5]. This may be related to depletion of ''F. prausnitzii'' in the microbiome, as ''F. prausnitzii'' has also shown to aid in serotonin restoration in the gut [16]. Serotonin is a neurotransmitter, which when reduced to low levels, has been tied to mood-related disorders. | ||
==References== | ==References== | ||
[https://msystems.asm.org/content/msys/2/1/e00164-16.full.pdf 1. Asnicar, F., Manara, S., Zolfo, M., Truong, D.T., Scholz, M., Armanini, F., Ferretti, P., Gorfer, V., Pedrotti, A., Tett, A., Segata, N. (2017). Studying vertical microbiome transmission from mothers to infants by stain-level metagenomic profiling. American Society for Microbiology. 2(1)] | [https://msystems.asm.org/content/msys/2/1/e00164-16.full.pdf 1. Asnicar, F., Manara, S., Zolfo, M., Truong, D.T., Scholz, M., Armanini, F., Ferretti, P., Gorfer, V., Pedrotti, A., Tett, A., Segata, N. (2017). Studying vertical microbiome transmission from mothers to infants by stain-level metagenomic profiling. American Society for Microbiology. 2(1)] | ||
[https://www.ncbi.nlm.nih.gov/nuccore/NZ_CP022479 2. Benevides, L.J., Martin, R., Miquel, S., Bridonneau, C., Robert, V., Hudault, S., Chain, F., Berteau, O., Azevedo, V., Soares, S.C., Chatel, J.M., Sokol, H., Bermudez-Humaran, L.G., Thomas, M. and Langella, P. (2016). Faecalibacterium prausnitzii A2-165 chromosome, complete genome. National Center for Biotechnology Information (NCBI).] | [https://www.ncbi.nlm.nih.gov/nuccore/NZ_CP022479 2. Benevides, L.J., Martin, R., Miquel, S., Bridonneau, C., Robert, V., Hudault, S., Chain, F., Berteau, O., Azevedo, V., Soares, S.C., Chatel, J.M., Sokol, H., Bermudez-Humaran, L.G., Thomas, M. and Langella, P. (2016). ''Faecalibacterium prausnitzii'' A2-165 chromosome, complete genome. National Center for Biotechnology Information (NCBI).] | ||
[https://ib.bioninja.com.au/options/untitled/b1-microbiology-organisms/gram-staining.html 3. BioNinja. (2020). Gram Staining.] | [https://ib.bioninja.com.au/options/untitled/b1-microbiology-organisms/gram-staining.html 3. BioNinja. (2020). Gram Staining.] | ||
[https://www.microbiologyresearch.org/docserver/fulltext/ijsem/24/2/ijs-24-2-225.pdf?expires=1587489967&id=id&accname=guest&checksum=3AB4F548BD838788ECB11693CA0A1CD5 4. Cato, E.P., Salmon, C.W., Moore, W.E.C. (1974). Fusobacterium prausnitzii (Hauduroy et al.) Moore and Holdeman: Emended Description and Designation of Neotype Strain. International Journal of Systemic Bacteriology. 24 (2) 225-229.] | [https://www.microbiologyresearch.org/docserver/fulltext/ijsem/24/2/ijs-24-2-225.pdf?expires=1587489967&id=id&accname=guest&checksum=3AB4F548BD838788ECB11693CA0A1CD5 4. Cato, E.P., Salmon, C.W., Moore, W.E.C. (1974). ''Fusobacterium prausnitzii'' (Hauduroy et al.) Moore and Holdeman: Emended Description and Designation of Neotype Strain. International Journal of Systemic Bacteriology. 24 (2) 225-229.] | ||
[https://www.crohnscolitisfoundation.org/mental-health/depression-anxiety 5. Crohn’s and Colitis Foundation. (2020). Depression and Anxiety.] | [https://www.crohnscolitisfoundation.org/mental-health/depression-anxiety 5. Crohn’s and Colitis Foundation. (2020). Depression and Anxiety.] | ||
[https://www.microbiologyresearch.org/docserver/fulltext/ijsem/52/6/0522141a.pdf?expires=1587474833&id=id&accname=guest&checksum=8EFE8BA8548FDD5BBEAADAB68C163CD3 6. Duncan, S.H., Hold, G.L., Harmsen, H.J.M., Stewart, C.S., Flint, H.J. (2002). Growth Requirements and Fermentation Products of Fusobacterium Prausnitzii, and a Proposal to Reclassify | [https://www.microbiologyresearch.org/docserver/fulltext/ijsem/52/6/0522141a.pdf?expires=1587474833&id=id&accname=guest&checksum=8EFE8BA8548FDD5BBEAADAB68C163CD3 6. Duncan, S.H., Hold, G.L., Harmsen, H.J.M., Stewart, C.S., Flint, H.J. (2002). Growth Requirements and Fermentation Products of ''Fusobacterium Prausnitzii'', and a Proposal to Reclassify it as ''Faecalibacterium Prausnitzii'' Gen. Nov., Comb. Nov. International Journal of Systematic and Evolutionary Microbiology. 52, 2141-2146.] | ||
[https://bmcgenomics.biomedcentral.com/track/pdf/10.1186/s12864-018-5313-6 7. Fitzgerald, C.B., Shkoporov, A.N., Sutton, T.D.S., Chaplin, A.V., Velayudhan, V., Ross, R.P., Hill, C. (2018). Comparative analysis of Faecalibacterium prausnitzii genomes shows a high level of genome plasticity and warrants separation into new species-level taxa. BMC Genomics. 19, 931.] | [https://bmcgenomics.biomedcentral.com/track/pdf/10.1186/s12864-018-5313-6 7. Fitzgerald, C.B., Shkoporov, A.N., Sutton, T.D.S., Chaplin, A.V., Velayudhan, V., Ross, R.P., Hill, C. (2018). Comparative analysis of ''Faecalibacterium prausnitzii'' genomes shows a high level of genome plasticity and warrants separation into new species-level taxa. BMC Genomics. 19, 931.] | ||
[https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0145485&type=printable 8. Foditsch, C., Pereira, R.V.V., Ganda, E.K., Gomez, M.S., Marques, E.C., Santin, T., Bicalho, R.C. (2015). Oral administration of Faecalibacterium prausnitzii decreased the incidence of severe diarrhea and related mortality rate and increased weight gain in pre-weaned dairy heifers. PLOS Journals.] | [https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0145485&type=printable 8. Foditsch, C., Pereira, R.V.V., Ganda, E.K., Gomez, M.S., Marques, E.C., Santin, T., Bicalho, R.C. (2015). Oral administration of ''Faecalibacterium prausnitzii'' decreased the incidence of severe diarrhea and related mortality rate and increased weight gain in pre-weaned dairy heifers. PLOS Journals.] | ||
[https://www.dairyherd.com/article/probiotic-supplement-could-improve-calf-health 9. Hanson, M. (2019). Probiotic Supplement Could Improve Calf Health.] | [https://www.dairyherd.com/article/probiotic-supplement-could-improve-calf-health 9. Hanson, M. (2019). Probiotic Supplement Could Improve Calf Health.] | ||
[https://jb.asm.org/content/jb/196/18/3289.full.pdf 10. Heinken, A., Khan, M.T., Paglia, G., Rodionov, D.A., Harmsen, H.J.M., Thiele, I. (2014). Functional Metabolic Map of Faecalibacterium prausnitzii, a beneficial human gut microbe. Journal of Bacteriology. 96 (18) 3289-3302.] | [https://jb.asm.org/content/jb/196/18/3289.full.pdf 10. Heinken, A., Khan, M.T., Paglia, G., Rodionov, D.A., Harmsen, H.J.M., Thiele, I. (2014). Functional Metabolic Map of ''Faecalibacterium prausnitzii'', a beneficial human gut microbe. Journal of Bacteriology. 96 (18) 3289-3302.] | ||
[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3400418/ 11. Khan, M.T., Duncan S.H., Stams, A.J.M., Maarten van Dijl, J., Flint, H.J., Harmsen, H.J.M. (2012). The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic-anoxic interphases. The ISME Journal. 6(8): 1578–1585.] | [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3400418/ 11. Khan, M.T., Duncan S.H., Stams, A.J.M., Maarten van Dijl, J., Flint, H.J., Harmsen, H.J.M. (2012). The gut anaerobe ''Faecalibacterium prausnitzii'' uses an extracellular electron shuttle to grow at oxic-anoxic interphases. The ISME Journal. 6(8): 1578–1585.] | ||
[https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0096097&type=printable 12. Khan, M.T., Maarten van Dijl, J., Harmsen, H.J.M. (2014). Antioxidants keep the potentially probiotic but highly oxygen-sensitive human gut bacterium Faecalibacterium prausnitzii alive at ambient air. PLOS Journals. 9(5).] | [https://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0096097&type=printable 12. Khan, M.T., Maarten van Dijl, J., Harmsen, H.J.M. (2014). Antioxidants keep the potentially probiotic but highly oxygen-sensitive human gut bacterium ''Faecalibacterium prausnitzii'' alive at ambient air. PLOS Journals. 9(5).] | ||
[https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-019-02174-1#citeas 13. Kumar, M., Garand, M. & Al Khodor, S. (2019). Integrating omics for a better understanding of Inflammatory Bowel Disease: a step towards personalized medicine. Journal of Translational Medicine. 17, 419.] | [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-019-02174-1#citeas 13. Kumar, M., Garand, M. & Al Khodor, S. (2019). Integrating omics for a better understanding of Inflammatory Bowel Disease: a step towards personalized medicine. Journal of Translational Medicine. 17, 419.] | ||
| Line 76: | Line 79: | ||
[https://science.sciencemag.org/content/sci/362/6418/eaat9076.full.pdf 14. Livak, Y., Byndloss, M.X., Bäumler, A.J. (2018). Colonocyte metabolism shapes the gut microbiota. Science. 362, 6418.] | [https://science.sciencemag.org/content/sci/362/6418/eaat9076.full.pdf 14. Livak, Y., Byndloss, M.X., Bäumler, A.J. (2018). Colonocyte metabolism shapes the gut microbiota. Science. 362, 6418.] | ||
[https://aem.asm.org/content/aem/78/2/420.full.pdf 15. Lopez-Siles, M. Khan, M.T., Duncan, S.H., Harmsen, H.J.M., Garcia-Gil, L.J., Flint, H.J. (2012). Cultured Representatives of Two Major Phylogroups of Human Colonic Faecalibacterium prausnitzii Can Utilize Pectin, Uronic Acids, and Host-Derived Substrates for Growth. Applied and Environmental Microbiology. 78, 2, pp.420-428.] | [https://aem.asm.org/content/aem/78/2/420.full.pdf 15. Lopez-Siles, M. Khan, M.T., Duncan, S.H., Harmsen, H.J.M., Garcia-Gil, L.J., Flint, H.J. (2012). Cultured Representatives of Two Major Phylogroups of Human Colonic ''Faecalibacterium prausnitzii'' Can Utilize Pectin, Uronic Acids, and Host-Derived Substrates for Growth. Applied and Environmental Microbiology. 78, 2, pp.420-428.] | ||
[https://www.nature.com/articles/ismej2016176.pdf 16. Lopez-Siles, M., Duncan, H.S., Garcia-Gil, L.J., Martinez-Medina, M. (2017). Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. Nature – The International Society for Microbial Ecology (ISME) Journal. 11, pp. 841-852.] | [https://www.nature.com/articles/ismej2016176.pdf 16. Lopez-Siles, M., Duncan, H.S., Garcia-Gil, L.J., Martinez-Medina, M. (2017). ''Faecalibacterium prausnitzii'': from microbiology to diagnostics and prognostics. Nature – The International Society for Microbial Ecology (ISME) Journal. 11, pp. 841-852.] | ||
[https://onlinelibrary.wiley.com/doi/epdf/10.1111/cmi.12805 17. Maier, E., Anderson, R.C., Altermann, E., Roy, N.C. (2018). Live Faecalibacterium prausnitzii induces greater TLR2 and TLR2/6 activation than the dead bacterium in an apical anaerobic co-culture system. Wiley Online Library: Cellular Microbiology. 20, 12805.] | [https://onlinelibrary.wiley.com/doi/epdf/10.1111/cmi.12805 17. Maier, E., Anderson, R.C., Altermann, E., Roy, N.C. (2018). Live ''Faecalibacterium prausnitzii'' induces greater TLR2 and TLR2/6 activation than the dead bacterium in an apical anaerobic co-culture system. Wiley Online Library: Cellular Microbiology. 20, 12805.] | ||
[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5492426/ 18. Martin, R., Miquel, S., Benevides, L., Bridonneau, C., Robert, V., Hudault, S., Chain, F., Azevedo, V., Chatel, J.M., Sokol, H., Bermúdez-Humarán, L.G., Thomas, M., Langella, P. (2017). Functional Characterization of Novel Faecalibacterium prausnitzii Strains Isolated from Healthy Volunteers: A Step Forward in the Use of F. prausnitzii as a Next-Generation Probiotic. Frontiers in Microbiology.] | [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5492426/ 18. Martin, R., Miquel, S., Benevides, L., Bridonneau, C., Robert, V., Hudault, S., Chain, F., Azevedo, V., Chatel, J.M., Sokol, H., Bermúdez-Humarán, L.G., Thomas, M., Langella, P. (2017). Functional Characterization of Novel ''Faecalibacterium prausnitzii'' Strains Isolated from Healthy Volunteers: A Step Forward in the Use of ''F. prausnitzii'' as a Next-Generation Probiotic. Frontiers in Microbiology.] | ||
[https://www.frontiersin.org/articles/10.3389/fmicb.2018.00346/full 19. Martin, R., Bermúdez-Humarán, L.G., Langella, P. (2018). Searching for the Bacterial Effector: The example of the multi-skilled commensal bacterium Faecalibacterium prausnitzii. Frontiers in Microbiology.] | [https://www.frontiersin.org/articles/10.3389/fmicb.2018.00346/full 19. Martin, R., Bermúdez-Humarán, L.G., Langella, P. (2018). Searching for the Bacterial Effector: The example of the multi-skilled commensal bacterium ''Faecalibacterium prausnitzii''. Frontiers in Microbiology.] | ||
[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4464665/ 20. Mueller, N.T., Bakacs, E., Combellick, J., Grigoryan, Z., Dominguez-Bello, M.G. (2015). The infant microbiome development: mom matters. Trends in Molecular Medicine. 21(2) 109-117.] | [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4464665/ 20. Mueller, N.T., Bakacs, E., Combellick, J., Grigoryan, Z., Dominguez-Bello, M.G. (2015). The infant microbiome development: mom matters. Trends in Molecular Medicine. 21(2) 109-117.] | ||
| Line 90: | Line 93: | ||
[https://www.britannica.com/science/T-cell 21. Rogers, K. (2009). T cell. The Encyclopaedia Britannica.] | [https://www.britannica.com/science/T-cell 21. Rogers, K. (2009). T cell. The Encyclopaedia Britannica.] | ||
[https://www.nature.com/articles/srep18507.pdf 22. Rossi, O., van Berkel, L.A., Chain, F., Khan, M.T., Taverne, N., Sokol, H., Duncan, S.H., Flint, H.J., Harmsen, H.J.M., Langella, P., Samsom, J.N., Wells, J.M. (2016). Faecalibacterium prausnitzii A2-165 has a high capacity to induce IL-10 in human and murine dendritic cells and modulates T cell responses. Nature Scientific Reports. 6, 18507.] | [https://www.nature.com/articles/srep18507.pdf 22. Rossi, O., van Berkel, L.A., Chain, F., Khan, M.T., Taverne, N., Sokol, H., Duncan, S.H., Flint, H.J., Harmsen, H.J.M., Langella, P., Samsom, J.N., Wells, J.M. (2016). ''Faecalibacterium prausnitzii'' A2-165 has a high capacity to induce IL-10 in human and murine dendritic cells and modulates T cell responses. Nature Scientific Reports. 6, 18507.] | ||
[https://www.ncbi.nlm.nih.gov/books/NBK470553/ 23. Sizar, O., Unakal, C.G. (2019). Gram-Positive Bacteria. National Center for Biotechnology Information.] | [https://www.ncbi.nlm.nih.gov/books/NBK470553/ 23. Sizar, O., Unakal, C.G. (2019). Gram-Positive Bacteria. National Center for Biotechnology Information.] | ||
24. Sokol, H., Seksik, P., Furet, J.P., Firmesse, O., Nion-Larmurier, I., Beaugerie, L., Cosnes, J., Corthier, G., Marteau, P., Doré, J. (2009). Low Counts of Faecalibacterium prausnitzii in Colitis Microbiota. Wiley InterScience. | [https://s3.amazonaws.com/academia.edu.documents/45756932/Soko_H_Seksik_P_Furet_JP_et_al._Low_coun20160518-2104-14nuzxj.pdf?response-content-disposition=inline%3B%20filename%3DLow_counts_of_Faecalibacterium_prausnitz.pdf&X-Amz-Algorithm=AWS4-HMAC-SHA256&X-Amz-Credential=ASIATUSBJ6BAMASSPX4S%2F20200430%2Fus-east-1%2Fs3%2Faws4_request&X-Amz-Date=20200430T140457Z&X-Amz-Expires=3600&X-Amz-SignedHeaders=host&X-Amz-Security-Token=IQoJb3JpZ2luX2VjENz%2F%2F%2F%2F%2F%2F%2F%2F%2F%2FwEaCXVzLWVhc3QtMSJIMEYCIQDaiZUqbJzM%2FSNoCRePW3PHYu7k00Tv614nS4GyXkOfaQIhAIbHvuRXAVRqULZQzhpvU9ZbqUIP9OIg99wYYsKBBDfIKrQDCBUQABoMMjUwMzE4ODExMjAwIgz92D7t%2Fps8ndAOxMAqkQOSldpWQIGrBQalnDa6UAudtnPeL3rH%2Fl7cFORrRRhFNPSdF%2F4MrAzDqFsL%2BPwuHPzcDEA7FptrXQbadw6HUcpJic411NTe3kC5mfwSeUgCakeSjhRp1%2FR%2BMRhE1cUTKTQn6hMrMSJgw8%2B%2BBVGhf015tGsTpUMFfuLdzTVJG0uK%2BJddx8xiSVXDPbeOpXC%2B25OX8E4%2BElUGfgS2Hs2DhJurOlhCdzS4bYdlUI6OxWDe9hEshqB87EfHdBvYT1eHDA%2BA2e8p5GJ42S%2FZqfcVURW94wdJv4v65EUXq%2FTYWTBIPxoudyJ3Rc7T13iWsnyy7%2FnrQ7Ho2SB11XI3Z8EWyiQvnyYRDGm1tK98CHYr%2BqKWIQTVW8ukje%2FYAiteBNZ020XdGZ0LSygdbQzHM3BmoLyNcGDmssK8MdhQ1FMVVoPCt3DitBVQy8DFUoiu8g%2FymCdUS4Uua5hsRmWrrVZn4TBNT4rNNocuaD9ITlaSt9GmUrQtj0PhLoC5GarQBltj6eAD6zoP44q7kdw8OLt9eCnYgjCX%2Far1BTrqAU9EJIep3qpWLzKAtMslAQBChEju04hghVL%2FZc5Gc2Saqa5ocLRaOonntqA%2BSnks1k2vEqGsq%2FSPWZqntbRSVL65A0jbAmgMH4q1OdRbGxX3p9OCUEEPUQKZjnPdeiROZ3yCfibp%2FfyFMp1FYrZyBZUcS%2F5TwjstPFrXuMhsEhC6m574EGnSGVNrckyxbjMzqHt28RVSKrSgWoDVToaTj6RHlj3xr6ijyaV%2BUyWhGXVN7TA%2BQ9rz9P14EFA%2Bu%2BEfOdNhXkfjdPzItcWqFhtIUpsCGOS9Fa9E7OdZeTRiZBlSphFmGC4ryICayQ%3D%3D&X-Amz-Signature=5b3278fc113d25cd341f19e605fee2161a05c0e4bed761a0a87bb27a65719ee5 24. Sokol, H., Seksik, P., Furet, J.P., Firmesse, O., Nion-Larmurier, I., Beaugerie, L., Cosnes, J., Corthier, G., Marteau, P., Doré, J. (2009). Low Counts of ''Faecalibacterium prausnitzii'' in Colitis Microbiota. Wiley InterScience.] | ||
[https://www.pnas.org/content/pnas/105/43/16731.full.pdf 25. Sokol, H., Pigneur, B., Watterlot, L., Lakhdari, O., Bermúdez-Humarán, L.G., Gratadoux, J.J., Blugeon, S., Bridonneau, C., Furet, J.P., Corthier, G., Grangette, C., Vasquez, N., Pochart, P., Trugnan, G., Thomas, G., Blottière, H.M., Doré, J., Marteau, P., Seksik, P., Langella, P. (2008). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proceedings of the National Academy of Sciences.] | [https://www.pnas.org/content/pnas/105/43/16731.full.pdf 25. Sokol, H., Pigneur, B., Watterlot, L., Lakhdari, O., Bermúdez-Humarán, L.G., Gratadoux, J.J., Blugeon, S., Bridonneau, C., Furet, J.P., Corthier, G., Grangette, C., Vasquez, N., Pochart, P., Trugnan, G., Thomas, G., Blottière, H.M., Doré, J., Marteau, P., Seksik, P., Langella, P. (2008). ''Faecalibacterium prausnitzii'' is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proceedings of the National Academy of Sciences.] | ||
[https://www.scientificamerican.com/article/among-trillions-of-microbes-in-the-gut-a-few-are-special/?print=true 26. Velasques-Manoff, M. (2019). Among Trillions of Microbes in the Gut, Few are Special. Scientific American.] | [https://www.scientificamerican.com/article/among-trillions-of-microbes-in-the-gut-a-few-are-special/?print=true 26. Velasques-Manoff, M. (2019). Among Trillions of Microbes in the Gut, Few are Special. Scientific American.] | ||
[https://bmcbiol.biomedcentral.com/track/pdf/10.1186/1741-7007-11-61 27. Wrzosek, L., Miquel, S., Noordine, M., Bouet, S., Chevalier-Curt, M.J., Robert, V., Philippe, C., Bridonneau, C., Cherbuy, C., Robbe-Masselot, C., Langella, P., Thomas, M. (2013). Bacterioides thetaiotamicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biology. 11(61).] | [https://bmcbiol.biomedcentral.com/track/pdf/10.1186/1741-7007-11-61 27. Wrzosek, L., Miquel, S., Noordine, M., Bouet, S., Chevalier-Curt, M.J., Robert, V., Philippe, C., Bridonneau, C., Cherbuy, C., Robbe-Masselot, C., Langella, P., Thomas, M. (2013). ''Bacterioides thetaiotamicron'' and ''Faecalibacterium prausnitzii'' influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biology. 11(61).] | ||
[https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6036887/ 28. Wu, X., Wu, Y., He, L., Wu, L., Wang, X., Liu, Z. (2018). Effects of the intestinal microbial metabolite butyrate on the development of colorectal cancer. Journal of Cancer. 9(14): 2510–2517.] | [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6036887/ 28. Wu, X., Wu, Y., He, L., Wu, L., Wang, X., Liu, Z. (2018). Effects of the intestinal microbial metabolite butyrate on the development of colorectal cancer. Journal of Cancer. 9(14): 2510–2517.] | ||
[https://journals.plos.org/plosone/article/figure?id=10.1371/journal.pone.0123013.g001 29. "Phenotypic Characteristics of ''F. prausnitzii'' strain HTF-F and A2-165." Plos One Journal.] | |||
==Author== | ==Author== | ||
Latest revision as of 19:15, 30 April 2020
Classification
Domain: Bacteria
Phylum: Firmicutes
Class: Clostridia
Order: Clostridiales
Family: Ruminococcaceae [NCBI link to find]
Species
|
NCBI: Taxonomy |
Faecalibacterium prausnitzii
Description and Significance
Faecalibacterium prausnitzii is a rod-shaped, non-motile, non-spore forming, strictly anaerobic bacterium [4, 7]. Its optimal growth temperature is approximately 37 °C [4], which parallels the average temperature of mammals. F. prausnitzii is extremely oxygen sensitive. Studies have shown that when exposed to ambient air, F. prausnitzii loses its viability within approximately 2 minutes [11, 6]. These attributes have led to challenges regarding the cultivation and preservation of F. prausnitzii [12].
Faecalibacterium prausnitzii colonization in the colon is considered a notable biomarker for a healthy human gastrointestinal tract, and plays a key role in the maintenance of overall gut homeostasis. In the healthy human gut, F. prausnitzii comprises around 5% of the gut microbiota [7, 10, 16, 25], and has been detected at levels up to 15% [7]. Decreased abundance of F. prausnitzii has been linked to gut inflammation and several bowel diseases, including Crohn’s disease, ulcerative colitis, and colorectal cancer [7, 16, 24, 25].
Because of its significant role in gut homeostasis, F. prausnitzii strains are of high interest as a next-generation probiotic [18, 26]. Recent research has focused on molecules and strategies aimed at maintaining F. prausnitzii viable when exposed to varying levels of oxygen. There have been encouraging studies showing F. prausnitzii maintaining its viability for 24 hours under ambient air, by exploiting extracellular antioxidants such as riboflavin and cysteine, among others [12, 10]. These studies are part of an effort to make F. prausnitzii commercially viable as a biotherapeutic remedy and probiotic to help those suffering from bowel diseases [13, 16, 19]. Other researchers have also seen the potential of F. prausnitzii in the agricultural industry as a probiotic for cattle [8, 9].
Genome Structure
Faecalibacterium prausnitzii has two main phylogroups: Phylogroup I and Phylogroup II [16]. Within these phylogroups, there are different strains whose genomes have been documented in public databases "with varying levels of assembly and annotation quality" [7]. The genome size of Faecalibacterium prausnitzii exhibits variation, ranging from 2.68 million base pairs (Mbp) to 3.42 Mbp and having wide range of G-C content, varying from 54.9% to 63.0% [7]. Most F. prausnitzii genomes have been constructed from draft assemblies. However, one of the first strains to have a complete genome representation is F. prausnitzii strain A2-165 (Phylogroup II) [7]. This strain of F. prausnitzii has a circular genome containing 3.11 Mbp, 56.3% G-C content, 3,017 total genes, 2,790 coding genes, and 85 RNA genes [3].
In a study by Fitzgerald et al. (2018), 31 genomes of high-quality draft as well as complete genomes were used in a comparative genomics analysis to observe intraspecies diversity. The results revealed a high level of genome plasticity and a relatively low level of average nucleotide identity (ANI) between F. prausnitzii groups. Based on this and other observations, Fitzgerald et al. have proposed to separate Faecalibacterium prausnitzii into two new species-level taxa [7].
Cell Structure, Metabolism and Life Cycle
Faecalibacterium prausnitzii shows irregular staining, but typically stains like a Gram-negative bacteria [4]. However, F. prausnitzii exhibits dermis characteristics that resemble Gram-positive bacteria, such as those in clostridial cluster IV (Clostridium leptum group) [6, 17]. Typical Gram-negative bacteria have LPS (lipopolysaccharide) on their outer membranes. LPS proteins are ligands for the transmembrane protein, TLR4 (Toll-like receptor 4). These are not found on F. prausnitzii or other bacteria in clostridial cluster IV. This suggests that F. prausnitzii is one of the unique bacteria whose dermis structure does not necessarily align with its Gram-stain [17].
F. prausnitzii is an acetate-consuming and butyrate-producing bacterium [27, 16]. Although it is one of the most common bacteria found in the human colon, it has been revealed that F. prausnitzii needs the presence of B. thetaiotamicron, a primary acetate-producer, in order to colonize the colon [27]. F. prausnitzii produces the short-chain fatty acid (SCFA) butyrate as a product of fermenting indigestible fiber [11]. Butyrate serves as one of the primary energy sources for colonocytes (cells lining the colon) [10] and is a principal molecule that works through various signaling pathways to prevent inflammation, regulate cell proliferation (cancer chemopreventative activities) [22], and maintain the integrity of the colon epithelial mucosa [16, 18, 19].
Ecology and Pathogenesis
Faecalibacterium prausnitzii is found ubiquitously in the gastrointestinal tracts of both animal and human hosts, and acts as a symbiotic, commensal bacterium within its host [16, 19]. One of the ways in which F. prausnitzii helps maintain a healthy colon is by the production of butyrate via the fermentation of fiber [10, 11]. Butyrate has demonstrated to have various attributes that positively contribute to colonic wellbeing and maintenance [14, 28].
Butyrate acts as a primary metabolite that maintains a symbiotic relationship between F. prausnitzii and colonocytes [10]. As F. prausnitzii produces butyrate, colonocytes respond with increased metabolic activity and cell-sustaining maintenance. These cellular activities deplete oxygen from the lumen of the colon, creating a hypoxic environment for obligate anaerobes to dominate [14]. As a result, the fermentation of fiber into short-chain fatty acids, which are a primary source of colonocyte energy, continues in a symbiotic manner [10, 16]. Higher levels of oxygen in the colon deplete the integrity of the microbiome and result in increased abundance of facultative anaerobic bacteria [14]. This is a tell-tale sign of a gut in dysbiosis and leads to greater inflammation and chronic irritable bowel diseases [14]. Current research has also focused on identifying different Faecalibacterium prausnitzii strains in patients as a biomarker, in order to discriminate between different gut diseases, hoping to lead to improved diagnostics and treatment [16].
Faecalibacterium prausnitzii’s further promotes gut health by the production and promotion of anti-inflammatory molecules. These molecules include a proteinaceous “microbial anti-inflammatory molecule” (MAM) that can block the NF-κB pathway of inflammatory response [16]. Other anti-inflammatory attributes observed from F. prausnitzii include the ability to upregulate anti-inflammatory cytokines, such as IL-10, downregulatre pro-inflammatory molecules such as IL-12 and IFN-γ, and “reduce the severity of acute, chronic, and low-grade chemical-induced inflammation" [16, 22, 25]. F. prausnitzii has been shown to attenuate the severity of inflammation by inducing the production of tight-junction proteins found in the intestinal mucosa. This anti-inflammatory activity maintains the integrity of the gut lining and prevents leaky gut and further inflammation [16]. Finally, F. prausnitzii has been found to play a role in regulating proper proportionality of cell types in the gut epithelial lining, leading to proper cell maintenance functions (such as apoptosis) for the evasion of inflammation and cancerous cells [15].
Rates of depression and anxiety have been associated with inflammatory bowel diseases such as Crohn’s disease and ulcerative colitis [5]. This may be related to depletion of F. prausnitzii in the microbiome, as F. prausnitzii has also shown to aid in serotonin restoration in the gut [16]. Serotonin is a neurotransmitter, which when reduced to low levels, has been tied to mood-related disorders.
References
3. BioNinja. (2020). Gram Staining.
5. Crohn’s and Colitis Foundation. (2020). Depression and Anxiety.
9. Hanson, M. (2019). Probiotic Supplement Could Improve Calf Health.
21. Rogers, K. (2009). T cell. The Encyclopaedia Britannica.
29. "Phenotypic Characteristics of F. prausnitzii strain HTF-F and A2-165." Plos One Journal.
Author
Page authored by Monica Pinal, student of Prof. Jay Lennon at Indiana University.