Coronavirus Invasion mechanism: Difference between revisions
No edit summary |
|||
| (151 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
<!-- Do not edit this line-->{{Curated}} | <!-- Do not edit this line-->{{Curated}} | ||
<b>Lizzy Apunda | <b>By Lizzy Apunda | ||
==Introduction== | ==Introduction== | ||
[[ | [[File:CoronavirusImage.jpg|thumb|200px|left|Figure 1:</b> An image of Coronavirus Disease 2019 (COVID 19). <ref name=az>[https://microbewiki.kenyon.edu/index.php/Coronavirus_Invasion_mechanism= "Newsroom" <i>Centers for Disease Control and Prevention</i>(CDC) 25 February 2019. Web. 14 April. 2020.]</ref>]] | ||
[[File:Coronavirusphylo.JPG|thumb|450px|right|<b>Figure 2:</b> Phylogenetic tree of 28 coronaviruses with complete protein sequences of helicase.https://cmr.asm.org/content/20/4/660.short <ref name=ay>[https://cmr.asm.org/content/20/4/660.short=Cheng, V. C., Lau, S. K., Woo, P. C., & Yuen, K. Y. (2007). Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. <i>Clinical microbiology reviews<i>, 20(4), 660-694.]</ref>]] | |||
The | <b>Coronaviruses</b> are a large family consisting of enveloped, non-segmented, positive stranded RNA viruses that cause moderate to mild upper-respiratory tract, gastrointestinal, hepatic and central nervous system diseases. <ref name=aa> [https://core.ac.uk/download/pdf/82753182.pdf=Gallagher, T., & Buchmeier, M. (2001). Coronavirus Spike Proteins in Viral Entry and Pathogenesis. <i> Virology<i>, 279(2), 371-374. doi: 10.1006/viro.2000.0757]</ref> | ||
These viruses have a broad host range and infect both mammals (pigs, camels, bats, cats e.t.c) and avian species. Rare circumstances known as <b>spillover events</b> cause the viruses to jump to humans and cause disease <ref name=ab>[https://www.nih.gov/health-information/coronavirus= “Coronavirus” <i>National Institutes of Health<i> (COVID-19). (2020). Retrieved 8 April 2020.]</ref>.The virus primarily causes <b>upper respiratory tract infections</b> in humans and fowls and <b>enteric infections</b> in porcine and bovine <ref name=aa/> . Since 2013, porcine epidemic diarrhea coronavirus (PEDV) has killed 100% of infected piglets in America <ref name=ac>[https://www.annualreviews.org/doi/abs/10.1146/annurev-virology-110615-042301=Li, F. (2016). Structure, Function, and Evolution of Coronavirus Spike Proteins. <i> Annual Review Of Virology<i>, 3(1), 237-261. doi: 10.1146/annurev-virology-110615-042301]</ref>. This constituted 10% of America’s pig population. About four of the seven known coronaviruses only cause mild to moderate symptoms in infected individuals. Three of these, however, are capable of causing severe, even fatal, disease: Severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV) and Coronavirus 2019 (COVID-19) (<b>Figure 1</b>) <ref name=ag/>. SARS-CoV emerged in November 2002 and disappeared in 2004 after infecting 8000 people with a fatality rate of ~10% <ref name=ab>[https://www.nih.gov/health-information/coronavirus= “Coronavirus” <i>National Institutes of Health<i> (COVID-19). (2020). Retrieved 8 April 2020.]</ref>. The sudden disappearance was likely due to intensive contact tracing and care isolation measures <ref name=ab>[https://www.nih.gov/health-information/coronavirus= “Coronavirus” <i>National Institutes of Health<i> (COVID-19). (2020). Retrieved 8 April 2020.]</ref>. Since 2012, MERS-CoV has infected more than 1700 people, with a fatality rate of ~36% <ref name=ac/>. Coronaviruses adapt to new environments through mutation and recombination and as a result can alter host range and tissue tropism efficiently <ref name=ae>[https://www.nature.com/articles/nature02145=Li, W., Moore, M., Vasilieva, N., Sui, J., Wong, S., & Berne, M. et al. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. <i>Nature<i>, 426(6965), 450-454. doi: 10.1038/nature02145]</ref> | |||
. This means that the effects of coronaviruses on global health and economic stability are constant and long term. Therefore, it is crucial to study and understand the virology of coronaviruses <ref name=ag/>. | |||
Coronaviruses belong to the family<i> [[Coronaviridae]]</i> in the order <i>Nidovirales</i> <ref name=ac/>. These viruses have a viral genome of about 26-32 kilobases and can further be classified into four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. These genera were first determined by serology, and later by phylogenetic clustering <ref name=ad>[https://doi.org/10.1007/978-1-4939-2438-7_1=Fehr, A. R., & Perlman, S. (2015). Coronaviruses: an overview of their replication and pathogenesis. <i>Methods in molecular biology<i>, (Clifton, N.J.), 1282, 1–23.]</ref> | |||
. Alpha and beta coronaviruses infect mammals, gamma coronaviruses infect avian species, and delta coronaviruses infect both mammalian and avian species <ref name=ad/>. Examples of alpha coronaviruses include Human coronavirus (HCoV-NL63), porcine transmissible gastroenteritis coronavirus (TGEV), PEDV, and porcine respiratory coronavirus (PRCV) <ref name=ac/>. Examples of beta coronaviruses include SARS-CoV, 2019-nCoV, MERS-CoV, bat coronavirus HKU4, mouse hepatitis coronavirus (MHV), bovine coronavirus (BCoV), and human coronavirus OC43 <ref name=ac/>. The 2019-nCoV also an example of a betacoronavirus that is ancestral to human SARS-CoV and bat SARS-CoV <ref name=af>[https://jvi.asm.org/content/94/7/e00127-20?_ga=2.265409295.1611640120.1581636020-280187052.1580834497=Wan, Y., Shang, J., Graham, R., Baric, R., & Li, F. (2020). Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. <i>Journal Of Virology<i>, 94(7). doi: 10.1128/jvi.00127-20]</ref>. Examples of gamma- and delta coronaviruses include avian infectious bronchitis coronavirus (IBV) and porcine deltacoronavirus (PdCV), respectively <ref name=ad/>.(<b>Figure 2</b>) illustrates that all the differnet types of coronaviruses evolved from a common ancestor. | |||
== | ==Genome Structure and Organization== | ||
[[ | [[File:Corona4.JPG|thumb|200px|left|<b>Figure 3:</b>Genomic organization of representative α, β, and γ CoVs. An illustration of the MHV genome is depicted at the top. The expanded regions below show the structural and accessory proteins in the 3′ regions of the HCoV-229E, MHV, SARS-CoV, MERS-CoV and IBV.<ref name=ad>[https://doi.org/10.1007/978-1-4939-2438-7_1=Fehr, A. R., & Perlman, S. (2015). Coronaviruses: an overview of their replication and pathogenesis. <i>Methods in molecular biology<i>, (Clifton, N.J.), 1282, 1–23.]</ref>]] | ||
[[File:CoronaX.JPG|thumb|300px|Right|<b>Figure 4:</b>Genomic organization of representative α, β, and γ CoVs. An illustration of the anatomy of SARS-CoV 2.<ref name=ayy>[https://www.economist.com/briefing/2020/03/12/understanding-sars-cov-2-and-the-drugs-that-might-lessen-its-power. <i>The Economist<i>. Retrieved 20 April 2020.]</ref>]] | |||
Viruses in the <i>Nidovirales</i> order have exceptionally large genome sizes among all RNA viruses, with the largest genome size being 33.5 kilobases <ref name=ag>[https://www.sciencedirect.com/science/article/pii/S0166354213002222?casa_token=5tMd2PZzKiQAAAAA:NDAcVoGRdxTuFz5bmWTaXe1x6Qh2etREcJAm0BYa2nb3mCwYweV0BmVYXBTP2L9yg6pEhLzUMA=Li, F. (2013). Receptor recognition and cross-species infections of SARS coronavirus. <i>Antiviral Research<i>, 100(1), 246-254. doi: 10.1016/j.antiviral.2013.08.014]</ref>. Coronaviruses have a highly organized genome structure where the 5’ ends have a cap while the 3’ ends have a poly(A) tail and some accessory proteins (<b>Figure 3</b>). <ref name=ad/>. The 5’ ends also contain untranslated regions, stem loop structures, and a leader sequence required for RNA replication and transcription of the viral genome. These features enable the genome to act as an mRNA for the translation of the replicase protein, which encodes non-structural proteins <ref name=ad/>. Since coronaviruses are positive-stranded RNA viruses, they do not need to package their RNA-dependent RNA polymerase, since the ribosomes will translate the RNA immediately <ref name=ad/>. The genome is packed inside a helical capsid, which is common in negative-sense strand RNA and unusual in positive-sense strand RNA viruses (<b>Figure 4</b>). These viruses have spike projections that protrude from the surface in addition to four structural proteins: the Spike protein (S), the membrane protein (M), the envelope protein (E) and the nucleocapsid protein (N) <ref name=ad/>. The S protein uses an N-terminal signal sequence to mediate attachment to the host receptor <ref name=ad/>. The M protein exists as a dimer and contains three transmembrane domains. This protein is responsible for giving the virion its shape and promotes membrane curvature. The E protein is a transmembrane protein that has various functions: facilitates assembly and dispersion of the virus, and contains ion channel activity. In SARS-CoV, the ion channel activity is necessary for pathogenesis <ref name=ad/>. Phosphorylation in the N protein triggers a structural change that increases the affinity for viral DNA <ref name=ad/>. | |||
==Coronaviruses Spike Proteins== | |||
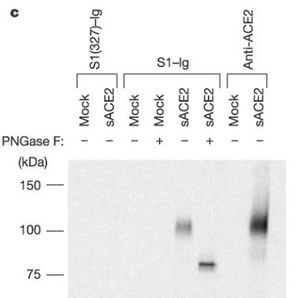
[[File:Coronajj.JPG|thumb|300px|left|<b>Figure 5: </b>Supernatants of radiolabelled 293T cells transfected with plasmid encoding soluble ACE2 or with vector alone (mock) were immunoprecipitated with S1(327)–Ig, S1–Ig, or an anti-ACE2 antibody. Immunoprecipitates were treated or not, as indicated, with PNGase F, and analysed by SDS–PAGE.<ref name=ae>[https://www.nature.com/articles/nature02145=Li, W., Moore, M., Vasilieva, N., Sui, J., Wong, S., & Berne, M. et al. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. <i>Nature<i>, 426(6965), 450-454. doi: 10.1038/nature02145]</ref>]] | |||
The coronavirus spike proteins associate with cellular receptors to facilitate infection of their target cells (Li et al, 2003). They consist of an ectodomain, a transmembrane anchor and a short intracellular tail <ref name=ac/>. The ectodomain contains the receptor binding subunit S1 that binds to the host’s cell surface during virus entry and a membrane fusion subunit S2, which fuses the host and viral membrane <ref name=ag/>. These processes are critical for the coronavirus infection cycle. <b>SARS-Cov</b> and <b>2019 nCoV</b> spike proteins share similarities of around 76-78%, while the receptor binding proteins share about 50-53% similarities <ref name=af>[https://jvi.asm.org/content/94/7/e00127-20?_ga=2.265409295.1611640120.1581636020-280187052.1580834497=Wan, Y., Shang, J., Graham, R., Baric, R., & Li, F. (2020). Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. <i>Journal Of Virology<i>, 94(7). doi: 10.1128/jvi.00127-20]</ref>. | |||
==Receptor Recognition== | |||
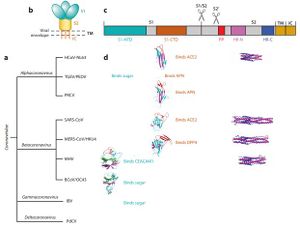
[[File:Corona3.JPG|thumb|300px|right|<b>Figure 6: </b>Introduction to coronaviruses and their spike proteins. (a) Classification of coronaviruses. | |||
(b) Schematic of the overall structure of prefusion coronavirus spikes. Shown are the receptor-binding subunit S1, the membrane-fusion subunit S2, the transmembrane anchor (TM), the intracellular tail (IC), and the viral envelope. (c) Schematic of the domain structure of coronavirus spikes, including the S1 N-terminal domain (S1-NTD), the S1 C-terminal domain (S1-CTD), the fusion peptide (FP), and heptad repeat regions N and C (HR-N and HR-C). Scissors indicate two proteolysis sites in coronavirus spikes. (d ) Summary of the structures and functions of coronavirus spikes.<ref name=ac>[https://www.annualreviews.org/doi/abs/10.1146/annurev-virology-110615-042301=Li, F. (2016). Structure, Function, and Evolution of Coronavirus Spike Proteins. <i> Annual Review Of Virology<i>, 3(1), 237-261. doi: 10.1146/annurev-virology-110615-042301]</ref>]] | |||
Coronaviruses have a rich diversity of receptor usage. They either utilize the <b>S1, N-terminal domain (S1-NTD)</b> or the <b>S1, C-terminal domain (S1-CTD) </b> as a receptor-binding domain <ref name=af>[https://jvi.asm.org/content/94/7/e00127-20?_ga=2.265409295.1611640120.1581636020-280187052.1580834497=Wan, Y., Shang, J., Graham, R., Baric, R., & Li, F. (2020). Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. <i>Journal Of Virology<i>, 94(7). doi: 10.1128/jvi.00127-20]</ref>. Coronavirus S1-NTDs bind sugar with the exception of the beta coronavirus MHV that binds a protein receptor <ref name=ag/>. The S1-CTDs recognize protein receptors <b>ACE2</b>, APN, and DPP4 <ref name=ad/>. Alpha coronaviruses such as the human coronavirus (HCoV-NL63) and beta coronaviruses such as SARS-CoV recognize the zinc peptidase angiotensin-converting enzyme 2 (ACE2) <ref name=ag/>. According to <b>Figure 5</b>, ACE2 is indeed a functional receptor for SARS-CoV as anti-ACE2 and not anti-ACE1 antibody blocked viral replication on Vero E6 cells. Other alpha coronaviruses TGEV, PEDV, and PRCV recognize the zinc peptidase, aminopeptidase N (APN) <ref name=ad/>. Comparably, other beta coronaviruses recognize different receptors: a serine peptidase, dipeptidyl peptidase 4 (DPP4) <ref name=ad/>. Alpha coronaviruses such as TGEV and PEDV, together with gamma coronavirus (IBV) use sugar as receptors or coreceptors <ref name=ad/>. | |||
(<b>Figure 6</b>) illustrates a summary of known receptors of gamma,beta, alpha and deltacoronaviruses. These receptors have other physiological functions aside from facilitating viral entry. The S1-CTD of the SARS-CoV exists as a core structure (five-stranded antiparallel β-sheet) and a receptor binding motif (RBM) <ref name=ad/>. The RBM includes a surface that binds the ACE2 receptor. SARS-Cov strains that were isolated from human patients and palm civets during the SARS epidemic showed differences in S1-CTD residues of the RBM region: <b>Asn479</b> and <b>Thr487</b> in human viral strains become <b>Lys479</b> and <b>Ser487</b> in civet viral strains, respectively <ref name=ac/>. Strains collected from the humans bound more tightly to the human ACE2 receptor than strains collected from the civets. These results were crucial in the study of cross-species transmissions of SARS-CoV <ref name=ag/>. Human ACE2 residues <b>Lys31</b> and <b>lys353</b> are virus hotspots with salt bridges and are instrumental in virus receptor binding. Protein residues that interact with these hotpots are under selective pressure to mutate <ref name=ad/>. Naturally selected viral mutations strengthen the structure of the hot spots, enhancing the binding affinity of S1-CTD for human ACE2 <ref name=ad/>. These mutations were responsible for the civet-to-human and human-to-human transmissions of the virus <ref name=ad/>. Rat, mouse and bat ACE2 protein residues are unable to bind to the SARS-CoV binding domain <ref name=ad/>. | |||
[ | The similarities in Receptor binding proteins and spike proteins of the 2019-nCoV and SARS-CoV suggest that the two may share the same receptor (ACE2) <ref name=af>[https://jvi.asm.org/content/94/7/e00127-20?_ga=2.265409295.1611640120.1581636020-280187052.1580834497=Wan, Y., Shang, J., Graham, R., Baric, R., & Li, F. (2020). Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. <i>Journal Of Virology<i>, 94(7). doi: 10.1128/jvi.00127-20]</ref>. Evidence that supports this is that the SARS-CoV receptor binding motifs do not have deletions or insertions. Nine of the 14 ACE2 residues in the RBM are fully conserved while 4 are partially conserved among human, bat and civet SARS-CoV and 2019-nCoV <ref name=af/>. Favorable interactions between residues and viral binding hot spots enhances viral binding of 2019-nCoV to human ACE2. The viral binding ACE2 residues of cats, ferrets, monkeys, pigs and orangutans have similar viral binding residues <ref name=ac/>. This explains why the 2019-nCov is able to recognize them. The diversity of bats makes it difficult to establish the ability of 2019-nCoV to bind to the ACE2 <ref name=ad/>. 2019-nCoV RBM recognizes the ACE2 sequence of the Rhinolophus sinicus bats <ref name=ac/>. | ||
==Coronavirus Replicase== | |||
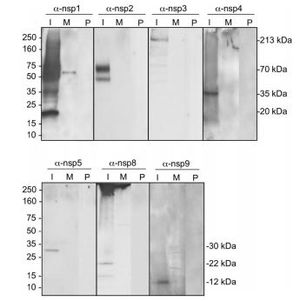
[[File:Corona7.JPG|thumb|300px|right|<b>Figure 8:</b> Identification of ORF1a SARS-CoV replicase proteins by immunoblotting ORF1a-encoded mature replicase proteins. SARS-CoV appears to encode only one polyprotein, which has similarity to PLP2 of other coronaviruses, and the protein identified in SARSCoV-infected cells was observed to be 20 kDa in mass.<ref name=ah>[https://jvi.asm.org/content/78/18/9977.short=Prentice, E., McAuliffe, J., Lu, X., Subbarao, K., & Denison, M. R. (2004). Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. <i>Journal of virology<i>, 78(18), 9977-9986.]</ref>]] | |||
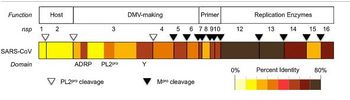
[[File:Coronajkl.JPG|thumb|350px|left|<b>Figure 7: </b> Conservation of the SARS-CoV replicase. Replicase subunits, or domains for nsp3, were color-coded according to percent identity between homologous proteins of SARS-CoV and MERS-CoV.<ref name=azz>[https://viralzone.expasy.org/. <i>Microbiology Class 2020 Google Slides<i>. Retrieved 20 April 2020.]</ref>]] | |||
[[ | |||
[[File:Coro.JPG|thumb|350px|left|<b>Figure 9: </b> Structures of the coronavirus replicase proteins nsp7, nsp8 and nsp9. Structures for the nsp7 and nsp8 heterodimers from SARS-CoV (2AHM; panel A) and FCoV (3UB0; panel B) are shown to illustrate the distinctive structures of these proteins.<ref name=azz>[https://viralzone.expasy.org/. <i>Microbiology Class 2020 Google Slides<i>. Retrieved 20 April 2020.]</ref>]] | |||
<i>Nidoviruses</i> possess a significant number of individual proteins compared to other plus strand viruses <ref name=ai>[https://link.springer.com/chapter/10.1007/3-540-26765-4_3=Ziebuhr, J. (2005). The coronavirus replicase. <i>In Coronavirus replication and reverse genetics<i>, (pp. 57-94). Springer, Berlin, Heidelberg.]</ref>. These extra proteins are necessary to produce a more useful replication and transcription system that increases the fidelity of RNA-dependent RNA synthesis. This process makes it possible to replace host factors that are needed by other viruses in an | |||
otherwise error prone RNA-dependent RNA synthesis <ref name=ah>[https://jvi.asm.org/content/78/18/9977.short=Prentice, E., McAuliffe, J., Lu, X., Subbarao, K., & Denison, M. R. (2004). Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. <i>Journal of virology<i>, 78(18), 9977-9986.]</ref>. It also assists the viruses to interact with the host cell and the immune system of the host animal. Gene expression in Coronaviruses begins with the translation of the replicase gene from the infectious genomic RNA <ref name=ai/>. | |||
The replicase gene consists of two large Open Reading Frames (ORF): ORF1a and ORF1b (<b>Figure 8</b>). The two types are located at the 5’ end and cover over two-thirds of the genome <ref name=aj>[https://www.microbiologyresearch.org/content/journal/jgv/10.1099/vir.0.80044-0=Hertzig, T., Scandella, E., Schelle, B., Ziebuhr, J., Siddell, S. G., Ludewig, B., & Thiel, V. (2004). Rapid identification of coronavirus replicase inhibitors using a selectable replicon RNA. <i>Journal of general virology<i>, 85(6), 1717-1725.]</ref>. The upstream ORF1a encodes a polyprotein, pp 1a, whereas a combination of the reading frames encode pp 1ab, which is translated as a ribosomal frameshift <ref name=aj/>. Some of the virally encoded proteinases such as papain-like and 3C like proteinases can process the coronavirus polyproteins pp 1a and pp 1b to form 15 to 16 end products and a number of intermediate products <ref name=aj/>. These non-structural proteins <b>(nsp)</b> assemble to form a fully functional replication and transcription complex in the cytoplasm of the infected cell: nsp1-nsp16 <ref name=jkl>[https://www.sciencedirect.com/science/article/pii/S0168170213004334=Neuman, B. W., Chamberlain, P., Bowden, F., & Joseph, J. (2014). Atlas of coronavirus replicase structure. Virus research, 194, 49-66.]</ref>. Nsp1 and nsp2 interfere with host defenses while nsp 3- 6 contain viral factors necessary to form viral replicative organelles in addition to two proteinases that process all the viral replicase proteins. Nsp 7- 11 (<b>Figure 9</b>) contain primer-making mechanisms while nsp 12- 16 contain the remaining RNA modifying enzymes required for replication. <b>Figure 7</b> depicts an illustration of SARS-CoV replicase <ref name=jkl/> | |||
. This complex provides a medium for replication and of coronavirus genomic RNA and transcription of multiple sub-genomic mRNA | |||
<ref name=aj/>. Research also suggests that the replicase gene may contribute to tropism and pathogenicity <ref name=ah/>. There is a significant number of conserved domains present in the replicase that are uncharacterized. These domains may provide potential targets for antiviral intervention examples including helicase,s proteases, and RNA-dependent RNA | |||
polymerase <ref name=aj/>. | |||
== | ==Discontinuous Transcription by Coronaviruses== | ||
[ | Whereas, plus and minus strands as large as the genome are generated continuously or processively by viral replicases, discontinuous transcription is a mechanism that is required to synthesize the minus-strand templates for the subgenomic mRNA <ref name=ak>[https://books.google.com/books?hl=en&lr=&id=Ja_kQu4lUMoC&oi=fnd&pg=PA1&dq=Coronavirus+replication+and+reverse+genetics+(Vol.+287).+Springer+Science+%26+Business+Media&ots=7OXGqEv_fl&sig=KyIlGHPG-JDu0Oazo4PGVwS6a3Q#v=onepage&q=Coronavirus%20replication%20and%20reverse%20genetics%20(Vol.%20287).%20Springer%20Science%20%26%20Business%20Media&f=false=Enjuanes, L. (Ed.). (2004). Coronavirus replication and reverse genetics (Vol. 287). Springer Science & Business Media.]</ref>. Studies on the group 2 coronavirus, Mouse Hepatitis Virus Strain (MHV-A59), revealed that all of the viral plus strands possessed the 1.7 kb sequence of RNA-7 as well as the poly (a) tract at their 3’ and 5’ ends <ref name=ak/>. In addition, the genome and all the sub-genomic mRNAs had similar leader sequences. Since this leader sequence is restricted to the 5’ end of the genome, scientists suggest that in viral RNA synthesis, there may be a way for the leader RNA to be joined to the body of mRNAs at the 3’ end of the genome <ref name=ak/>. The end of the leader sequence and before the ORF of the sub-genomic RNA, there exists a translation regulating/activating sequence (TRS- UCUAAAC) <ref name=ak>[https://books.google.com/books?hl=en&lr=&id=Ja_kQu4lUMoC&oi=fnd&pg=PA1&dq=Coronavirus+replication+and+reverse+genetics+(Vol.+287).+Springer+Science+%26+Business+Media&ots=7OXGqEv_fl&sig=KyIlGHPG-JDu0Oazo4PGVwS6a3Q#v=onepage&q=Coronavirus%20replication%20and%20reverse%20genetics%20(Vol.%20287).%20Springer%20Science%20%26%20Business%20Media&f=false=Enjuanes, L. (Ed.). (2004). Coronavirus replication and reverse genetics (Vol. 287). Springer Science & Business Media.]</ref>. This replication strategy occurs in <i>Arteriviridae</i> in the <i>Coronaviridae</i> family <ref name=ak/>. | ||
According to Enjuanes’ model of the 3’ discontinuous extension, viral polymerase begins transcription at the 3’ end of the genome (2004) and pauses after transcription of the TRS- UCUAAAC sequence. Every polymerase that gets to this point can either continue transcription or move to the 5’ end of the genome without copying the intervening sequences. This process is known as discontinuous transcription <ref name=ak/>. The coronaviruses polymerases may function in a way that is analogous to DNA dependent RNA polymerases where the polymerase is primarily associated with the growing strand <ref name=ak/>. Scientists suggest that a similar mechanism may exist for proofreading by RNA polymerase: the polymerase pauses, retracts and then excises nucleotides from the 3’ end <ref name=ak/>. Several gene products of the ORF1b of coronaviruses and SARS- CoV were identified to function in this manner, with added nuclease activity <ref name=ak/>. | |||
== | ==Assembly and Release== | ||
[[File:Coronazz.JPG|thumb|300px|left|<b>Figure 10: </b> An image illustrating the translation of RNA-dependent RNA polymerase and the creation of virus-generated vesicles "Virus factories".<ref name=azz>[https://viralzone.expasy.org/. <i>Microbiology Class 2020 Google Slides<i>. Retrieved 20 April 2020.]</ref>]] | |||
After replication and sub-genomic RNA synthesis, coronaviruses assemble intracellularly at membranes of the immediate compartment <ref name=ad>[https://doi.org/10.1007/978-1-4939-2438-7_1=Fehr, A. R., & Perlman, S. (2015). Coronaviruses: an overview of their replication and pathogenesis. <i>Methods in molecular biology<i>, (Clifton, N.J.), 1282, 1–23.]</ref>. This usually occurs between the endoplasmic reticulum (ER) and the golgi apparatus (<b>Figure 10</b>)<ref name=am>[https://www.embopress.org/doi/abs/10.1002/j.1460-2075.1996.tb00553.x=Vennema, H., Godeke, G. J., Rossen, J. W., Voorhout, W. F., Horzinek, M. C., Opstelten, D. J., & Rottier, P. J. (1996). Nucleocapsid‐independent assembly of coronavirus‐like particles by co‐expression of viral envelope protein genes. <i>The EMBO journal<i>, 15(8), 2020-2028.]</ref>. This process involves the viral structural proteins, S, E and M getting inserted into the ER where they move along the secretory pathway into the <b>ER-golgi intermediate compartment (ERGIC)</b> <ref name=am/> | |||
= | The helical nucleocapsids that are generated in the cytoplasm align at these membranes and mingle with the cytoplasmic domains of the viral membrane proteins <ref name=ad/>. At the ERGIC, the viral genomes that have been enclosed by the N protein bud into membranes of the ERGIC. These membranes contain viral structural proteins and form mature virions <ref name=ad/>. The virions are transported from the cell and into the <b>exocytic pathway</b> as they undergo various post assembly maturation processes including proteolytic and oligosaccharide processing <ref name=am/> The M protein is responsible for managing the majority of protein-protein interactions needed for the assembly of coronaviruses <ref name=ad/>. The co-expression of the M protein and the E protein is sufficient for the formation of virus-like particles. This suggests that the two proteins are required for the production of the coronavirus envelope <ref name=ad/>. The N protein enhances the formation of the particles while the S protein traffics to the ERGIC and interacts with the M protein enhancing incorporation of virions <ref name=ad/>. Researchers have suggested that the E protein may be required for inducing membrane curvature, altering the host secretory pathway or preventing the aggregation of the M protein <ref name=ad/>. The M protein binds to the nucleocapsids, an interaction that enhances the completion of <b>virion assembly</b>. In several coronaviruses, the S protein travels to the cell surface where it mediates cell-cell fusion between infected and adjacent uninfected cells <ref name=ad/>. This process causes the formation of giant, multinucleated cells that enables the virus to spread within an infected organism without detection or neutralization by antibodies <ref name=ad/>. | ||
==Pathogenesis== | |||
===Animal Coronaviruses=== | |||
Coronaviruses are responsible for causing a large number of diseases in animals especially livestock. Approximately 75% of emerging infectious diseases are of zoonotic origin <ref name=an>[https://pdfs.semanticscholar.org/0a6d/2d51211b4ab745f0af000920a700cecbf820.pdf=Saif, L. J. (2004). Animal coronaviruses: what can they teach us about the severe acute respiratory syndrome?. <i>Revue scientifique et technique-Office international des épizooties<i>, 23(2), 643-660.]</ref> | |||
.Transmissible Gastroenteritis Virus is a coronavirus that infects pigs by binding to the APN receptor <ref name=ao>[https://link.springer.com/chapter/10.1007/978-1-4899-1531-3_16=Enjuanes, L., & van der Zeijst, B. A. (1995). Molecular basis of transmissible gastroenteritis virus epidemiology. <i>In The coronaviridae<i> (pp. 337-376). Springer, Boston, MA.]</ref>. Porcine Epidemic Diarrhea Virus (PEDV) infects a pig’s intestinal cell lining and causes severe dehydration and diarrhea. Porcine hemagglutinating encephalomyelitis virus (PHEV) causes an enteric infection in pigs with the added possibility of infecting the nervous system <ref name=ad/>. Feline enteric coronavirus (FCoV) causes a mild, asymptomatic infection in domestic cats. This strain becomes virulent with persistent infection <ref name=ad/>. Bovine CoV, Rat CoV, and Infectious Bronchitis Virus (IBV) lead to the formation of mild to severe respiratory tract diseases in livestock, rats, and chickens respectively <ref name=ad/>. Murine hepatitis virus (MHV) infects mice and causes respiratory, enteric, hepatic and neurologic diseases. These infections have been used as model systems to study the effects of the coronavirus. Animal coronaviruses lead to high mortality and morbidity in livestock, which negatively impacts the economy. | |||
[[ | ===Human Coronaviruses=== | ||
[[File:Coronapic.JPG|thumb|300px|bottom|<b>Figure 11:</b> Summary of host range and cross-species infections of SARS-CoV. RBD mutations that overcame species barriers for the cross-species infections of SARS-CoV are in red and labeled as “+”. Residue changes in ACE2 that form species barriers and disfavor the cross-species infections of SARS-CoV are in green and labeled as “−”.<ref name=ag/>]] | |||
For a long period, coronaviruses were believed to only cause mild, respiratory tract infections in humans. The SARS-CoV was the first to debunk this theory. Betacoronaviruses such as <b>HCoV 229E</b> and the </b>HCoV OC43 were the first human coronaviruses to be identified <ref name=al>[https://doi.org/10.3390/v4113044=Geller, C., Varbanov, M., & Duval, R. E. (2012). Human coronaviruses: insights into environmental resistance and its influence on the development of new antiseptic strategies. <i>Viruses<i>, 4(11), 3044–3068.] </ref>. The two were responsible for upper and mild respiratory tract infections like the common cold <ref name=al/>. After the emergence of other coronaviruses, such as <b>NL63</b> in 2004, <b>HKU1</b> in 2005 and SARS-CoV in 2003, new studies have emerged characterizing HCoV. Research shows that human coronavirus infections mainly occur in the winter, with a relatively short incubation period <ref name=al/>. Coronaviruses can cause bronchitis, bronchiolitis or pneumonia <ref name=ad/>. These infections predominantly occur in weak patients i.e newborns/infants, the elderly and immunocompromised patients <ref name=al/>. The HCoVs are believed to cause digestive issues and necrotizing enterocolitis in newborns <ref name=al/>. Diarrhea and other gastrointestinal issues may accompany coronavirus infections <ref name=ad/>. Some HCoV OC43 infected patients exhibit neurological symptoms suggesting the possible involvement of the HCoV in the Central Nervous System. | |||
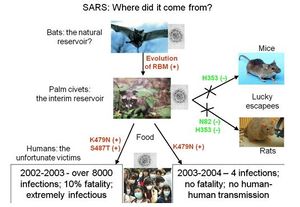
Highly pathogenic coronaviruses such as <b>SARS-CoV</b> and <b>Covid-19</b> affect a significant number of people in the world. SARS-CoV and Covid19 infections in humans cause fatigue, rigors, high fever, and tiredness <ref name=al/>. Covid19 patients also report having shortness of breath. A third of patients infected with SARS-CoV recover as clinical symptoms regress, however, some continue to have persistent pulmonary lesions <ref name=ad/>. Covid!9 has an even lower infection rate of around 1%. Respiratory insufficiency in both diseases cause respiratory failure, which is the most common cause of death among infected patients <ref name=al/>. The majority of patients infected with SARS-CoV develop watery diarrhea with active virus shedding for several weeks, which increases transmissibility <ref name=ad/>. The ability of coronaviruses to jump from one species to the next poses a risk to the human population <ref name=ad/>. For instance, the HCoV OC43 may have evolved from the bovine coronavirus and SARS-CoV is a zoonotic virus that crossed the species barrier (<b>Figure 11</b>)<ref name=al/>. | |||
==Vaccines and Therapy== | |||
There is currently no treatment or vaccine to fight HCoVs. The major strategies employed by healthcare professionals includes helping patients manage their symptoms until they recover. Multi-organ failure, respiratory failure and septic shock is the leading cause of death in Covid-19 patients. | |||
<br><br>Authored for BIOL 238 Microbiology, taught by [mailto:slonczewski@kenyon.edu Joan Slonczewski], 2018, [http://www.kenyon.edu/index.xml Kenyon College]. | <br><br>Authored for BIOL 238 Microbiology, taught by [mailto:slonczewski@kenyon.edu Joan Slonczewski], 2018, [http://www.kenyon.edu/index.xml Kenyon College]. | ||
Latest revision as of 19:11, 15 May 2020
By Lizzy Apunda
Introduction
Coronaviruses are a large family consisting of enveloped, non-segmented, positive stranded RNA viruses that cause moderate to mild upper-respiratory tract, gastrointestinal, hepatic and central nervous system diseases. [3] These viruses have a broad host range and infect both mammals (pigs, camels, bats, cats e.t.c) and avian species. Rare circumstances known as spillover events cause the viruses to jump to humans and cause disease [4].The virus primarily causes upper respiratory tract infections in humans and fowls and enteric infections in porcine and bovine [3] . Since 2013, porcine epidemic diarrhea coronavirus (PEDV) has killed 100% of infected piglets in America [5]. This constituted 10% of America’s pig population. About four of the seven known coronaviruses only cause mild to moderate symptoms in infected individuals. Three of these, however, are capable of causing severe, even fatal, disease: Severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV) and Coronavirus 2019 (COVID-19) (Figure 1) [6]. SARS-CoV emerged in November 2002 and disappeared in 2004 after infecting 8000 people with a fatality rate of ~10% [4]. The sudden disappearance was likely due to intensive contact tracing and care isolation measures [4]. Since 2012, MERS-CoV has infected more than 1700 people, with a fatality rate of ~36% [5]. Coronaviruses adapt to new environments through mutation and recombination and as a result can alter host range and tissue tropism efficiently [7] . This means that the effects of coronaviruses on global health and economic stability are constant and long term. Therefore, it is crucial to study and understand the virology of coronaviruses [6].
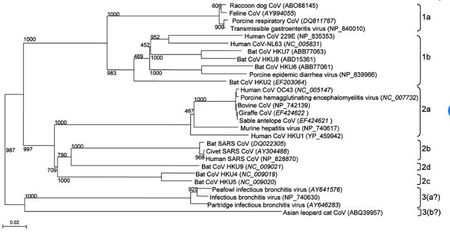
Coronaviruses belong to the family Coronaviridae in the order Nidovirales [5]. These viruses have a viral genome of about 26-32 kilobases and can further be classified into four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. These genera were first determined by serology, and later by phylogenetic clustering [8] . Alpha and beta coronaviruses infect mammals, gamma coronaviruses infect avian species, and delta coronaviruses infect both mammalian and avian species [8]. Examples of alpha coronaviruses include Human coronavirus (HCoV-NL63), porcine transmissible gastroenteritis coronavirus (TGEV), PEDV, and porcine respiratory coronavirus (PRCV) [5]. Examples of beta coronaviruses include SARS-CoV, 2019-nCoV, MERS-CoV, bat coronavirus HKU4, mouse hepatitis coronavirus (MHV), bovine coronavirus (BCoV), and human coronavirus OC43 [5]. The 2019-nCoV also an example of a betacoronavirus that is ancestral to human SARS-CoV and bat SARS-CoV [9]. Examples of gamma- and delta coronaviruses include avian infectious bronchitis coronavirus (IBV) and porcine deltacoronavirus (PdCV), respectively [8].(Figure 2) illustrates that all the differnet types of coronaviruses evolved from a common ancestor.
Genome Structure and Organization
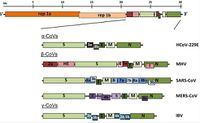
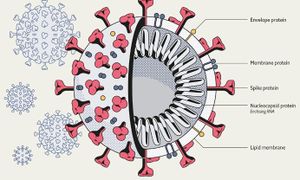
Viruses in the Nidovirales order have exceptionally large genome sizes among all RNA viruses, with the largest genome size being 33.5 kilobases [6]. Coronaviruses have a highly organized genome structure where the 5’ ends have a cap while the 3’ ends have a poly(A) tail and some accessory proteins (Figure 3). [8]. The 5’ ends also contain untranslated regions, stem loop structures, and a leader sequence required for RNA replication and transcription of the viral genome. These features enable the genome to act as an mRNA for the translation of the replicase protein, which encodes non-structural proteins [8]. Since coronaviruses are positive-stranded RNA viruses, they do not need to package their RNA-dependent RNA polymerase, since the ribosomes will translate the RNA immediately [8]. The genome is packed inside a helical capsid, which is common in negative-sense strand RNA and unusual in positive-sense strand RNA viruses (Figure 4). These viruses have spike projections that protrude from the surface in addition to four structural proteins: the Spike protein (S), the membrane protein (M), the envelope protein (E) and the nucleocapsid protein (N) [8]. The S protein uses an N-terminal signal sequence to mediate attachment to the host receptor [8]. The M protein exists as a dimer and contains three transmembrane domains. This protein is responsible for giving the virion its shape and promotes membrane curvature. The E protein is a transmembrane protein that has various functions: facilitates assembly and dispersion of the virus, and contains ion channel activity. In SARS-CoV, the ion channel activity is necessary for pathogenesis [8]. Phosphorylation in the N protein triggers a structural change that increases the affinity for viral DNA [8].
The coronavirus spike proteins associate with cellular receptors to facilitate infection of their target cells (Li et al, 2003). They consist of an ectodomain, a transmembrane anchor and a short intracellular tail [5]. The ectodomain contains the receptor binding subunit S1 that binds to the host’s cell surface during virus entry and a membrane fusion subunit S2, which fuses the host and viral membrane [6]. These processes are critical for the coronavirus infection cycle. SARS-Cov and 2019 nCoV spike proteins share similarities of around 76-78%, while the receptor binding proteins share about 50-53% similarities [9].
Receptor Recognition
Coronaviruses have a rich diversity of receptor usage. They either utilize the S1, N-terminal domain (S1-NTD) or the S1, C-terminal domain (S1-CTD) as a receptor-binding domain [9]. Coronavirus S1-NTDs bind sugar with the exception of the beta coronavirus MHV that binds a protein receptor [6]. The S1-CTDs recognize protein receptors ACE2, APN, and DPP4 [8]. Alpha coronaviruses such as the human coronavirus (HCoV-NL63) and beta coronaviruses such as SARS-CoV recognize the zinc peptidase angiotensin-converting enzyme 2 (ACE2) [6]. According to Figure 5, ACE2 is indeed a functional receptor for SARS-CoV as anti-ACE2 and not anti-ACE1 antibody blocked viral replication on Vero E6 cells. Other alpha coronaviruses TGEV, PEDV, and PRCV recognize the zinc peptidase, aminopeptidase N (APN) [8]. Comparably, other beta coronaviruses recognize different receptors: a serine peptidase, dipeptidyl peptidase 4 (DPP4) [8]. Alpha coronaviruses such as TGEV and PEDV, together with gamma coronavirus (IBV) use sugar as receptors or coreceptors [8].
(Figure 6) illustrates a summary of known receptors of gamma,beta, alpha and deltacoronaviruses. These receptors have other physiological functions aside from facilitating viral entry. The S1-CTD of the SARS-CoV exists as a core structure (five-stranded antiparallel β-sheet) and a receptor binding motif (RBM) [8]. The RBM includes a surface that binds the ACE2 receptor. SARS-Cov strains that were isolated from human patients and palm civets during the SARS epidemic showed differences in S1-CTD residues of the RBM region: Asn479 and Thr487 in human viral strains become Lys479 and Ser487 in civet viral strains, respectively [5]. Strains collected from the humans bound more tightly to the human ACE2 receptor than strains collected from the civets. These results were crucial in the study of cross-species transmissions of SARS-CoV [6]. Human ACE2 residues Lys31 and lys353 are virus hotspots with salt bridges and are instrumental in virus receptor binding. Protein residues that interact with these hotpots are under selective pressure to mutate [8]. Naturally selected viral mutations strengthen the structure of the hot spots, enhancing the binding affinity of S1-CTD for human ACE2 [8]. These mutations were responsible for the civet-to-human and human-to-human transmissions of the virus [8]. Rat, mouse and bat ACE2 protein residues are unable to bind to the SARS-CoV binding domain [8]. The similarities in Receptor binding proteins and spike proteins of the 2019-nCoV and SARS-CoV suggest that the two may share the same receptor (ACE2) [9]. Evidence that supports this is that the SARS-CoV receptor binding motifs do not have deletions or insertions. Nine of the 14 ACE2 residues in the RBM are fully conserved while 4 are partially conserved among human, bat and civet SARS-CoV and 2019-nCoV [9]. Favorable interactions between residues and viral binding hot spots enhances viral binding of 2019-nCoV to human ACE2. The viral binding ACE2 residues of cats, ferrets, monkeys, pigs and orangutans have similar viral binding residues [5]. This explains why the 2019-nCov is able to recognize them. The diversity of bats makes it difficult to establish the ability of 2019-nCoV to bind to the ACE2 [8]. 2019-nCoV RBM recognizes the ACE2 sequence of the Rhinolophus sinicus bats [5].

Nidoviruses possess a significant number of individual proteins compared to other plus strand viruses [13]. These extra proteins are necessary to produce a more useful replication and transcription system that increases the fidelity of RNA-dependent RNA synthesis. This process makes it possible to replace host factors that are needed by other viruses in an otherwise error prone RNA-dependent RNA synthesis [11]. It also assists the viruses to interact with the host cell and the immune system of the host animal. Gene expression in Coronaviruses begins with the translation of the replicase gene from the infectious genomic RNA [13].
The replicase gene consists of two large Open Reading Frames (ORF): ORF1a and ORF1b (Figure 8). The two types are located at the 5’ end and cover over two-thirds of the genome [14]. The upstream ORF1a encodes a polyprotein, pp 1a, whereas a combination of the reading frames encode pp 1ab, which is translated as a ribosomal frameshift [14]. Some of the virally encoded proteinases such as papain-like and 3C like proteinases can process the coronavirus polyproteins pp 1a and pp 1b to form 15 to 16 end products and a number of intermediate products [14]. These non-structural proteins (nsp) assemble to form a fully functional replication and transcription complex in the cytoplasm of the infected cell: nsp1-nsp16 [15]. Nsp1 and nsp2 interfere with host defenses while nsp 3- 6 contain viral factors necessary to form viral replicative organelles in addition to two proteinases that process all the viral replicase proteins. Nsp 7- 11 (Figure 9) contain primer-making mechanisms while nsp 12- 16 contain the remaining RNA modifying enzymes required for replication. Figure 7 depicts an illustration of SARS-CoV replicase [15] . This complex provides a medium for replication and of coronavirus genomic RNA and transcription of multiple sub-genomic mRNA [14]. Research also suggests that the replicase gene may contribute to tropism and pathogenicity [11]. There is a significant number of conserved domains present in the replicase that are uncharacterized. These domains may provide potential targets for antiviral intervention examples including helicase,s proteases, and RNA-dependent RNA polymerase [14].
Whereas, plus and minus strands as large as the genome are generated continuously or processively by viral replicases, discontinuous transcription is a mechanism that is required to synthesize the minus-strand templates for the subgenomic mRNA [16]. Studies on the group 2 coronavirus, Mouse Hepatitis Virus Strain (MHV-A59), revealed that all of the viral plus strands possessed the 1.7 kb sequence of RNA-7 as well as the poly (a) tract at their 3’ and 5’ ends [16]. In addition, the genome and all the sub-genomic mRNAs had similar leader sequences. Since this leader sequence is restricted to the 5’ end of the genome, scientists suggest that in viral RNA synthesis, there may be a way for the leader RNA to be joined to the body of mRNAs at the 3’ end of the genome [16]. The end of the leader sequence and before the ORF of the sub-genomic RNA, there exists a translation regulating/activating sequence (TRS- UCUAAAC) [16]. This replication strategy occurs in Arteriviridae in the Coronaviridae family [16].
According to Enjuanes’ model of the 3’ discontinuous extension, viral polymerase begins transcription at the 3’ end of the genome (2004) and pauses after transcription of the TRS- UCUAAAC sequence. Every polymerase that gets to this point can either continue transcription or move to the 5’ end of the genome without copying the intervening sequences. This process is known as discontinuous transcription [16]. The coronaviruses polymerases may function in a way that is analogous to DNA dependent RNA polymerases where the polymerase is primarily associated with the growing strand [16]. Scientists suggest that a similar mechanism may exist for proofreading by RNA polymerase: the polymerase pauses, retracts and then excises nucleotides from the 3’ end [16]. Several gene products of the ORF1b of coronaviruses and SARS- CoV were identified to function in this manner, with added nuclease activity [16].
Assembly and Release
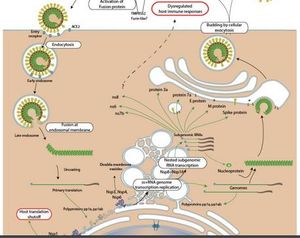
After replication and sub-genomic RNA synthesis, coronaviruses assemble intracellularly at membranes of the immediate compartment [8]. This usually occurs between the endoplasmic reticulum (ER) and the golgi apparatus (Figure 10)[17]. This process involves the viral structural proteins, S, E and M getting inserted into the ER where they move along the secretory pathway into the ER-golgi intermediate compartment (ERGIC) [17]
The helical nucleocapsids that are generated in the cytoplasm align at these membranes and mingle with the cytoplasmic domains of the viral membrane proteins [8]. At the ERGIC, the viral genomes that have been enclosed by the N protein bud into membranes of the ERGIC. These membranes contain viral structural proteins and form mature virions [8]. The virions are transported from the cell and into the exocytic pathway as they undergo various post assembly maturation processes including proteolytic and oligosaccharide processing [17] The M protein is responsible for managing the majority of protein-protein interactions needed for the assembly of coronaviruses [8]. The co-expression of the M protein and the E protein is sufficient for the formation of virus-like particles. This suggests that the two proteins are required for the production of the coronavirus envelope [8]. The N protein enhances the formation of the particles while the S protein traffics to the ERGIC and interacts with the M protein enhancing incorporation of virions [8]. Researchers have suggested that the E protein may be required for inducing membrane curvature, altering the host secretory pathway or preventing the aggregation of the M protein [8]. The M protein binds to the nucleocapsids, an interaction that enhances the completion of virion assembly. In several coronaviruses, the S protein travels to the cell surface where it mediates cell-cell fusion between infected and adjacent uninfected cells [8]. This process causes the formation of giant, multinucleated cells that enables the virus to spread within an infected organism without detection or neutralization by antibodies [8].
Pathogenesis
Coronaviruses are responsible for causing a large number of diseases in animals especially livestock. Approximately 75% of emerging infectious diseases are of zoonotic origin [18] .Transmissible Gastroenteritis Virus is a coronavirus that infects pigs by binding to the APN receptor [19]. Porcine Epidemic Diarrhea Virus (PEDV) infects a pig’s intestinal cell lining and causes severe dehydration and diarrhea. Porcine hemagglutinating encephalomyelitis virus (PHEV) causes an enteric infection in pigs with the added possibility of infecting the nervous system [8]. Feline enteric coronavirus (FCoV) causes a mild, asymptomatic infection in domestic cats. This strain becomes virulent with persistent infection [8]. Bovine CoV, Rat CoV, and Infectious Bronchitis Virus (IBV) lead to the formation of mild to severe respiratory tract diseases in livestock, rats, and chickens respectively [8]. Murine hepatitis virus (MHV) infects mice and causes respiratory, enteric, hepatic and neurologic diseases. These infections have been used as model systems to study the effects of the coronavirus. Animal coronaviruses lead to high mortality and morbidity in livestock, which negatively impacts the economy.
For a long period, coronaviruses were believed to only cause mild, respiratory tract infections in humans. The SARS-CoV was the first to debunk this theory. Betacoronaviruses such as HCoV 229E and the HCoV OC43 were the first human coronaviruses to be identified [20]. The two were responsible for upper and mild respiratory tract infections like the common cold [20]. After the emergence of other coronaviruses, such as NL63 in 2004, HKU1 in 2005 and SARS-CoV in 2003, new studies have emerged characterizing HCoV. Research shows that human coronavirus infections mainly occur in the winter, with a relatively short incubation period [20]. Coronaviruses can cause bronchitis, bronchiolitis or pneumonia [8]. These infections predominantly occur in weak patients i.e newborns/infants, the elderly and immunocompromised patients [20]. The HCoVs are believed to cause digestive issues and necrotizing enterocolitis in newborns [20]. Diarrhea and other gastrointestinal issues may accompany coronavirus infections [8]. Some HCoV OC43 infected patients exhibit neurological symptoms suggesting the possible involvement of the HCoV in the Central Nervous System.
Highly pathogenic coronaviruses such as SARS-CoV and Covid-19 affect a significant number of people in the world. SARS-CoV and Covid19 infections in humans cause fatigue, rigors, high fever, and tiredness [20]. Covid19 patients also report having shortness of breath. A third of patients infected with SARS-CoV recover as clinical symptoms regress, however, some continue to have persistent pulmonary lesions [8]. Covid!9 has an even lower infection rate of around 1%. Respiratory insufficiency in both diseases cause respiratory failure, which is the most common cause of death among infected patients [20]. The majority of patients infected with SARS-CoV develop watery diarrhea with active virus shedding for several weeks, which increases transmissibility [8]. The ability of coronaviruses to jump from one species to the next poses a risk to the human population [8]. For instance, the HCoV OC43 may have evolved from the bovine coronavirus and SARS-CoV is a zoonotic virus that crossed the species barrier (Figure 11)[20].
Vaccines and Therapy
There is currently no treatment or vaccine to fight HCoVs. The major strategies employed by healthcare professionals includes helping patients manage their symptoms until they recover. Multi-organ failure, respiratory failure and septic shock is the leading cause of death in Covid-19 patients.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2018, Kenyon College.
- ↑ "Newsroom" Centers for Disease Control and Prevention(CDC) 25 February 2019. Web. 14 April. 2020.
- ↑ V. C., Lau, S. K., Woo, P. C., & Yuen, K. Y. (2007). Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clinical microbiology reviews, 20(4), 660-694.
- ↑ 3.0 3.1 T., & Buchmeier, M. (2001). Coronavirus Spike Proteins in Viral Entry and Pathogenesis. Virology, 279(2), 371-374. doi: 10.1006/viro.2000.0757
- ↑ 4.0 4.1 4.2 “Coronavirus” National Institutes of Health (COVID-19). (2020). Retrieved 8 April 2020.
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 5.8 5.9 F. (2016). Structure, Function, and Evolution of Coronavirus Spike Proteins. Annual Review Of Virology, 3(1), 237-261. doi: 10.1146/annurev-virology-110615-042301
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 6.6 6.7 F. (2013). Receptor recognition and cross-species infections of SARS coronavirus. Antiviral Research, 100(1), 246-254. doi: 10.1016/j.antiviral.2013.08.014
- ↑ 7.0 7.1 W., Moore, M., Vasilieva, N., Sui, J., Wong, S., & Berne, M. et al. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature, 426(6965), 450-454. doi: 10.1038/nature02145
- ↑ 8.00 8.01 8.02 8.03 8.04 8.05 8.06 8.07 8.08 8.09 8.10 8.11 8.12 8.13 8.14 8.15 8.16 8.17 8.18 8.19 8.20 8.21 8.22 8.23 8.24 8.25 8.26 8.27 8.28 8.29 8.30 8.31 8.32 8.33 8.34 8.35 8.36 8.37 A. R., & Perlman, S. (2015). Coronaviruses: an overview of their replication and pathogenesis. Methods in molecular biology, (Clifton, N.J.), 1282, 1–23.
- ↑ 9.0 9.1 9.2 9.3 9.4 Y., Shang, J., Graham, R., Baric, R., & Li, F. (2020). Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. Journal Of Virology, 94(7). doi: 10.1128/jvi.00127-20
- ↑ The Economist. Retrieved 20 April 2020.
- ↑ 11.0 11.1 11.2 E., McAuliffe, J., Lu, X., Subbarao, K., & Denison, M. R. (2004). Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. Journal of virology, 78(18), 9977-9986.
- ↑ 12.0 12.1 12.2 Microbiology Class 2020 Google Slides. Retrieved 20 April 2020.
- ↑ 13.0 13.1 J. (2005). The coronavirus replicase. In Coronavirus replication and reverse genetics, (pp. 57-94). Springer, Berlin, Heidelberg.
- ↑ 14.0 14.1 14.2 14.3 14.4 T., Scandella, E., Schelle, B., Ziebuhr, J., Siddell, S. G., Ludewig, B., & Thiel, V. (2004). Rapid identification of coronavirus replicase inhibitors using a selectable replicon RNA. Journal of general virology, 85(6), 1717-1725.
- ↑ 15.0 15.1 B. W., Chamberlain, P., Bowden, F., & Joseph, J. (2014). Atlas of coronavirus replicase structure. Virus research, 194, 49-66.
- ↑ 16.0 16.1 16.2 16.3 16.4 16.5 16.6 16.7 16.8 L. (Ed.). (2004). Coronavirus replication and reverse genetics (Vol. 287). Springer Science & Business Media.
- ↑ 17.0 17.1 17.2 H., Godeke, G. J., Rossen, J. W., Voorhout, W. F., Horzinek, M. C., Opstelten, D. J., & Rottier, P. J. (1996). Nucleocapsid‐independent assembly of coronavirus‐like particles by co‐expression of viral envelope protein genes. The EMBO journal, 15(8), 2020-2028.
- ↑ L. J. (2004). Animal coronaviruses: what can they teach us about the severe acute respiratory syndrome?. Revue scientifique et technique-Office international des épizooties, 23(2), 643-660.
- ↑ L., & van der Zeijst, B. A. (1995). Molecular basis of transmissible gastroenteritis virus epidemiology. In The coronaviridae (pp. 337-376). Springer, Boston, MA.
- ↑ 20.0 20.1 20.2 20.3 20.4 20.5 20.6 20.7 C., Varbanov, M., & Duval, R. E. (2012). Human coronaviruses: insights into environmental resistance and its influence on the development of new antiseptic strategies. Viruses, 4(11), 3044–3068.
