Deamination model of hypermutation: Difference between revisions
No edit summary |
No edit summary |
||
| Line 9: | Line 9: | ||
==AID== | ==AID== | ||
[[Image:AID deamination.jpg|thumb|400px|right| Activation Induced Cytidine Deaminase(AID) in activated B cell https://www.sciencedirect.com/science/article/pii/S0022283612008984]]The AID protein itself is 198 amino acids long and is encoded by the AICDA region of the DNA. AID belongs to the APOBEC-1-related protein family which includes other DNA editing molecules. The molecule contains a cytosine deaminase domain as well as a leucine rich region around the c terminus, the latter of which is likely involved in a protein-protein interaction. Within B cells, AID initiates the processes of both SHM and class switch recombination(CSR), although different parts of the protein are involved in each process. The C terminus of the AID protein is associated with CSR. | [[Image:AID deamination.jpg|thumb|400px|right| Activation Induced Cytidine Deaminase(AID) in activated B cell https://www.sciencedirect.com/science/article/pii/S0022283612008984]]The AID protein itself is 198 amino acids long and is encoded by the AICDA region of the DNA. AID belongs to the APOBEC-1-related protein family which includes other DNA editing molecules. The molecule contains a cytosine deaminase domain as well as a leucine rich region around the c terminus, the latter of which is likely involved in a protein-protein interaction. Within B cells, AID initiates the processes of both SHM and class switch recombination(CSR), although different parts of the protein are involved in each process. The C terminus of the AID protein is associated with CSR. The expression of C-terminal mutations in the AICDA gene lead to decreased levels of CSR as the level of SHM remains constant<ref>Durandy, Anne, Sophie Peron, Nadine Taubenheim, and Alain Fischer. "Activation‐induced cytidine deaminase: structure–function relationship as based on the study of mutants." Human mutation 27, no. 12 (2006): 1185-1191.https://onlinelibrary.wiley.com/doi/epdf/10.1002/humu.20414</ref>. Similarly, mutations of the N terminus led to the defective induction of SHM suggesting that the N terminus of AID plays some sort of role in initiating SHM. | ||
Revision as of 17:43, 9 December 2021
BIOL 116 Class 2020 Jump to: navigation, search
Introduction
Deamination is a proposed mechanism of somatic hypermutation(SHM) which results in point mutations within the Immunoglobulin variable region(IgV) gene. Deamination relies chiefly on the use of activation-induced deaminase(AID), a protein which converts cytidine(cytosine bound to ribose) to uridine(uracil bound to ribose). This process results in a miss match of bases and therefore initiates DNA repair. However, the repair process uses mutagenic mechanisms, such as error-prone DNA polymerases, which result in the creation of mutations in and rearrangement of IgV genes. These mutations code for a varying expression of the gene itself resulting in an antibody with either a higher or lower binding affinity. After hypermutation, higher binding affinity is selected for during affinity maturation resulting in a more effective antibody. Mutagenic processes like those induced by AID must be tightly regulated due to the disease related and oncogenic risks of mutating DNA. Therefore, the AID protein is controlled by a complex network of regulatory factors which are vital to the safety of SHM.
AID
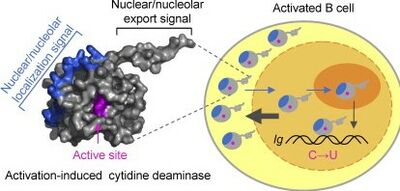
The AID protein itself is 198 amino acids long and is encoded by the AICDA region of the DNA. AID belongs to the APOBEC-1-related protein family which includes other DNA editing molecules. The molecule contains a cytosine deaminase domain as well as a leucine rich region around the c terminus, the latter of which is likely involved in a protein-protein interaction. Within B cells, AID initiates the processes of both SHM and class switch recombination(CSR), although different parts of the protein are involved in each process. The C terminus of the AID protein is associated with CSR. The expression of C-terminal mutations in the AICDA gene lead to decreased levels of CSR as the level of SHM remains constant[1]. Similarly, mutations of the N terminus led to the defective induction of SHM suggesting that the N terminus of AID plays some sort of role in initiating SHM.
Mechanism and regulation
AID carries a potential tumorigenic nature when mistargeted on to non-Ig loci. AID has been shown to promote B cell leukemia, lymphoma, and myeloma. Therefore the regulation of AID is as critical as the mechanism itself in terms of protecting the body. Regulation of AID starts with the AICDA gene and the transcription factors which manage its expression. AICDA is composed of 4 regions which include binding sites for at least 19 transcription factors. While transcription factors like NFκB, STAT6 and C/EBP bind to enhancer regions which promote transcription, transcription factors like PAX5 and E2A bind to and inactivate repressor regions of the gene therefore also promoting transcription. Most of these transcription factors are only upregulated during B cell activation, subjugating incidents of hypermutation as a response to B cell activation only. High transcription levels of AID are not desired in non-differentiating B cells. Excess levels of AID are linked to more frequent targeting of non IgV region genes inducing undesired mutations which may be associated with cancers. As a result, the level of AID is also heavily regulated on a post-transcription level using miRNAs. Two main miRNAs(miR-155 and miR-181b) target transcripts of the AICDA gene. Both miRNAs bind to the 3’ untranslated region of AID mRNA inducing a mechanism that ultimately results in lowered levels of SHM and CSR. Despite targeting the same sites, miR-181b differs in its expression pattern from miR-155. miR-181b levels are highest in unstimulated B cells and its levels decrease heavily as a result of activation. However, miR-181 levels gradually recover after activation. It should be noted that miR-181 and miR-155 have multiple target molecules besides AID mRNA and therefore the expression patterns of the two molecules may not be entirely based on the regulation of AID levels. AID protein is further regulated by the cell using protein localization. The concentration of AID in the nucleus is regulated by multiple mechanisms, ultimately resulting in a higher concentration of AID in the cytoplasm than in the nucleus. One mechanism involves an anchor sequence on the C-terminal region of AID which hitches the protein to the cytoplasm. Another mechanism involves the active transport of AID into the nucleus via the protein’s N-terminal NLS(nuclear localization signal). Finally, AID is transported out of the nucleus via the protein’s NES(nuclear export signal) which binds to the CRM1 export protein carrying the molecule into the cytoplasm. Concentration of AID is also regulated by the half life of the molecule within and outside of the nucleus. While AID has a half life of around 8 hours in the cytoplasm, nuclear AID has a half life of only about 2.5 hours due to higher levels of polyubiquitination by proteasomes. The reason being that high levels of AID degradation in the nucleus prevent non Ig-targeting by keeping AID concentrations low. AID is phosphorylated at multiple sites, inducing the molecule to associate with RPA(replication protein A) which guides AID to both the V and S regions of the immunoglobulin gene. However the mechanism by which AID binds specifically to the V region over other loci is still unknown. RPA can only bind to single stranded DNA segments and therefore relies on either transcription bubbles or single stranded bubbles formed by other means to bind AID to its DNA substrate. AID may move along with the transcription bubble as it moves down the DNA. Once bound, AID deaminates Cytidine, converting the molecule into uridine. This creates a mismatch which is subsequently identified by the MSH2-MSH6 heterodimer. This molecule guides DNA Polη to the mismatch site. Polη is a type of translesional DNA polymerase, a class of polymerase which is specifically used to repair damaged DNA(et al). However, during SHM, Polη repairs deoxyuracil mismatches with low fidelity often inserting different base pairs from the original and as a result causing point mutations(Et al). While Polη plays the most significant role in the erroneous repair of SHM, it is not the only repair molecule involved. Proteins like Polζ and Rev1 also include mutations, Rev1 for example causes Guanine:cytosine inversions.
Effect of microbiota on AID
The human body hosts a vast ecosystem of microorganisms termed the human microbiome. This microbiome is chiefly composed of bacteria and lives most prominently in the human gastrointestinal tract[2] . While the majority of these organisms are harmless and or beneficial, some present a risk to the health of the human host. Some of these microbes, like Helicobacter Pylori, may cause tumorigenesis via the damage and mutation of DNA. H. Pylori causes this damage, in part, through inducing aberrant expression of AID in host cells(https://www.cell.com/trends/cancer/fulltext/S2405-8033(20)30058-3#back-bb0035). While when functioning properly, AID is only present in B cells during activation, H pylori causes the expression of AID in epithelial cells within the gastrointestinal tract. H pylori activates NF-κB transcription factor within host cells through the secretion of bacterial virulence factors. NF-κB is in part responsible for activating the transcription of the AID gene leading researchers to believe that H. Pylori uses the activation of NF-κB to promote the expression of AID in epithelial cells(https://www.sciencedirect.com/science/article/pii/S0952791510000865#bib21). This not only demonstrates the potential danger of the human microbiome but also the inherent dilemma of AID as a DNA mutator and the issues as well as benefits that arise from such a molecule.
Open the BIOL 116 Class 2020 template page in "edit."
Copy ALL the text from the edit window.
Then go to YOUR OWN page; edit tab. PASTE into your own page, and edit.
Electron micrograph of the Ebola Zaire virus. This was the first photo ever taken of the virus, on 10/13/1976. By Dr. F.A. Murphy, now at U.C. Davis, then at the CDC.[1].
At right is a sample image insertion. It works for any image uploaded anywhere to MicrobeWiki. The insertion code consists of: Double brackets: [[ Filename: PHIL_1181_lores.jpg Thumbnail status: |thumb| Pixel size: |300px| Placement on page: |right| Legend/credit: Electron micrograph of the Ebola Zaire virus. This was the first photo ever taken of the virus, on 10/13/1976. By Dr. F.A. Murphy, now at U.C. Davis, then at the CDC. Closed double brackets: ]]
Other examples: Bold Italic Subscript: H2O Superscript: Fe3+
I don't know
Section 1 Genetics Include some current research, with at least one image.
Sample citations: [1] [2]
A citation code consists of a hyperlinked reference within "ref" begin and end codes.
References
- ↑ Durandy, Anne, Sophie Peron, Nadine Taubenheim, and Alain Fischer. "Activation‐induced cytidine deaminase: structure–function relationship as based on the study of mutants." Human mutation 27, no. 12 (2006): 1185-1191.https://onlinelibrary.wiley.com/doi/epdf/10.1002/humu.20414
- ↑ Thomas, Ryan M., and Christian Jobin. "The microbiome and cancer: is the ‘oncobiome’mirage real?." Trends in cancer 1, no. 1 (2015): 24-35.https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4642279/
Edited by Luke Smallwood, student of Joan Slonczewski for BIOL 116 Information in Living Systems, 2020, Kenyon College.
