Cucurbit yellow stunting disorder virus (CYSDV): Difference between revisions
| Line 38: | Line 38: | ||
<br> | <br> | ||
<br> | <br> | ||
1. Virus penetrates into the host cell. The CYSDV is transfered into the host through the saliva of its vector, <i>Bemicia tabaci</i>, the whitefly, which feeds on the leaves of the host plant. The host range of the CYSDV is limited to members of the family <i>Cucurbitaceae</i>. | 1. Virus penetrates into the host cell. Like other plant viruses, the CYSDV requires mechanical transmission, such as abrasions of the leaf surface by the feeding of an insect, to infect their hosts. The CYSDV is transfered into the host through the saliva of its vector, <i>Bemicia tabaci</i>, the whitefly, which feeds on the leaves of the host plant. The host range of the CYSDV is limited to members of the family <i>Cucurbitaceae</i>. | ||
<br> | <br> | ||
2. Uncoating, and release of the viral genomic RNA into the cytoplasm. The precise process of uncoating the CYSDV genome remains unclear. | 2. Uncoating, and release of the viral genomic RNA into the cytoplasm. The precise process of uncoating the CYSDV genome remains unclear. | ||
| Line 48: | Line 48: | ||
5. Formation of new virus particles. The virion body is assembled by the major capsid protein (CP) and the tail by the minor capsid protein (CPm) [3] [6]. | 5. Formation of new virus particles. The virion body is assembled by the major capsid protein (CP) and the tail by the minor capsid protein (CPm) [3] [6]. | ||
<br> | <br> | ||
6. Release of progeny virions. | 6. Release of progeny virions. The CYSDV virions spread to unifected cells by traveling through plasmodesmata. | ||
==Viral Ecology & Pathology== | ==Viral Ecology & Pathology== | ||
Revision as of 03:38, 10 September 2010
A Viral Biorealm page on the family Cucurbit yellow stunting disorder virus (CYSDV)

Baltimore Classification
Group IV: (+) sense single-stranded RNA viruses
Higher order categories
Order: Unassigned
Family: Closteroviridae
Genus: Crinivirus
Description and Significance
The Cucurbit yellow stunting disorder virus, CYSDV, can infect members of the family Cucurbitaceae, including all types of melons, summer and winter squash, pumpkins, gourds, and cucumbers [1]. The CYSDV symptoms develop first in older leaves and mimic water stress. Intervenial chlorosis, a yellowing between the veins, streaks the leaves. Eventually the entire leaf becomes yellow except for the veins, which remain green [2]. In certain varieties, small green spots may develop on the leaves as well. As the plant’s internal transport system breaks down, it begins to drop older leaves in attempt to preserve itself. Without enough leaves, the plant’s strength dwindles and it no longer can support or nourish its fruit. As a result, the fruits are smaller, not as sweet, and don’t ship or store as well [1]. Consequently, the CYSDV has significant economic implications on national and international agriculture. However, control is difficult because there is no chemical or biological control suitable to fight the virus. Increasing water and fertilizer, and early season insecticide application (to reduce the virus’s vector population) may help, but these remedies compromise the producer’s ability to grow a sustainable crop and are incredibly costly [1].
Genome Structure
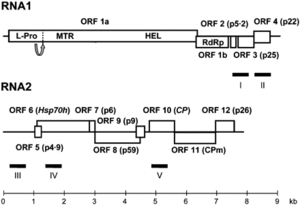
The CYSDV genome consists of two molecules of (+) sense ssRNA designated RNA1 and RNA2.
Although over seventy isolates of the CYSDV have been identified around the world, in countries including Spain, Jordan, Turkey, Lebanon, Saudi Arabia, and North America, two genetically distinct subpopulations have been identified. The two subpopulations are the so-called Eastern subpopulation, composed of the Saudia Arabian isolates and the Western subpopulation, containing the rest of the CYSDV isolates [2] [4]. The genomes of many of the isolates have yet to be sequenced. However, the complete nucleotide (nt) sequences of genomic RNAs 1 and 2 of for the Spanish isolate CYSDV-AlLM are known.
RNA1 is 9123 nt long and contains at least five open reading frames (ORFs) [5]. RNA2 is 7976 nt long and contains the gene array distinctive to the family Closteroviridae,
characterized by ORFs encoding a heat shock protein 70 homologue (HSP70h), two proteins of unknown function (58 and p9), the major coat protein (CP) and a divergent copy of the coat protein (CPm) [2] [4] [5].
Virion Structure of CYSDV
The CYSDV particles are non-enveloped, flexuous rods 750-800 nm in length. The virion body is assembled by the major capsid protein (CP) and the tail by the minor capsid protein (CPm). The virus ecapsidates 2 molecules of (+)sense ssRNA, designated RNA1 and RNA2 [2].
Reproductive Cycle of CYSDV in a Host Cell
Little is specifically known about the CYSDV reproductive cycle, but members of the Family Closteroviridae generally replicate in this way:
1. Virus penetrates into the host cell. Like other plant viruses, the CYSDV requires mechanical transmission, such as abrasions of the leaf surface by the feeding of an insect, to infect their hosts. The CYSDV is transfered into the host through the saliva of its vector, Bemicia tabaci, the whitefly, which feeds on the leaves of the host plant. The host range of the CYSDV is limited to members of the family Cucurbitaceae.
2. Uncoating, and release of the viral genomic RNA into the cytoplasm. The precise process of uncoating the CYSDV genome remains unclear.
3. The viral RNA is translated in the cytoplasm by host ribosomes to make RNA-dependent RNA polymerase.
4. The RNA-dependent RNA polymerase uses the viral RNA template to make (-) strand RNA (complementary to the [+] strand) to form a double-stranded RNA intermediate. The (-)strand RNA serves as a template for viral mRNAs, and progeny genomic RNA.
5. Formation of new virus particles. The virion body is assembled by the major capsid protein (CP) and the tail by the minor capsid protein (CPm) [3] [6].
6. Release of progeny virions. The CYSDV virions spread to unifected cells by traveling through plasmodesmata.
Viral Ecology & Pathology

References
[1] McGinley, Susan. 2010. "New virus attacks melons, cucumbers, and squash." Western Farm Press. http://westernfarmpress.com/new-virus-attacks-melons-cucumbers-and-squash-0. Date accessed: 6 Sept. 2010.
[2] Auguilar, Juan M. et al. Resistance to Cucurbit yellow stunting disorder virus in Cucumber. Plant Diseases. 90.5 (2006): 583.
[3] "Viral Zone: Closteroviridae". ExPASY Proteomics Server. http://www.expasy.org/viralzone/all_by_species/34.html.
[4] Rubio, Luis et al. Geographically distant isolates of the crinivirus Cucurbit yellow stunting disorder virus show very low genetic diversity in the coat protein gene. Journal of General Virology. 82 (2001): 929.
[5] Augilar, Juan M. et al. Further variability within the genus Crinivirus, as revealed by determination of the complete RNA genome sequence of Cucurbit yellow stunting disorder virus. Journal of General Virology. 84 (2003): 2555.
[6] Slonczewski, J.L., J.W. Foster. “Chapter 6: Virus Structure and Function.” Microbiology: An Evolving Science (2010): 193-207.
Page authored by Sally Wilson for BIOL 375 Virology, September 2010
