Hepatitis B Virus X gene in Hepatocellular Carcinoma: Difference between revisions
Engelbrechte (talk | contribs) No edit summary |
Engelbrechte (talk | contribs) No edit summary |
||
| Line 2: | Line 2: | ||
{{Viral Biorealm Family}} | {{Viral Biorealm Family}} | ||
Hepatocellular carcinoma (HCC) is among the most frequently diagnosed cancers worldwide. For over forty years physicians and researchers have observed an association between HBV infection and development of HCC. Of the seven HBV genes, hepatitis b virus x protein (HBx, 17.5 kDa) is the smallest and required extensive efforts to characterize; its elusiveness is highlighted by its designation “X” [1]. The protein functions as a transcriptional transactivator of both viral and cellular transcripts, in addition inducing cytoplasmic signal transduction pathways [1]. Interestingly, HBx was a more potent inducer of transcription upon deletion of its N-terminal 1 to 50-amino-acid fragment, suggesting this domain may function as a negative regulatory element [1]. | |||
[[Image:hep b.jpg|thumb|400px|right|<b>Figure 1</b>. text. [cite].]] | [[Image:hep b.jpg|thumb|400px|right|<b>Figure 1</b>. text. [cite].]] | ||
| Line 14: | Line 13: | ||
hep b infection, HCC | hep b infection, HCC | ||
== | ==Virus Overview== | ||
Hepatitis B virus is most commonly transmitted by contact with infected blood or body fluids. The virus may also be vertically transmitted during childbirth. The infection localizes in the liver and has been linked to the development of several diseases including cirrhosis and hepatocellular carcinoma. The hepatitis B vaccine contains hepatitis B surface antigen (HBsAg). The doses of the vaccine yield a sufficient adaptive immune response capable of | |||
In vitro, HBx binds eukaryotic basal transcription factors essential for proper RNAP II-DNA interactions and promoter escape, including TATA-binding protein (TBP), TFIIB, TFIIH, and the RNAP subunit RPB5 [ | |||
==Hepatitis B Virus Genome== | |||
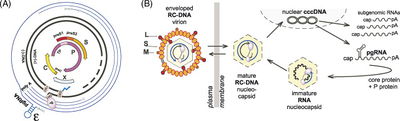
The hepadnaviral genome is packaged as relaxed circular (RC), partially double-stranded DNA about 3.2 kb in length. After viral DNA enters the nucleus, the partially double-stranded viral DNA is repaired and the (+) DNA strand is extended, rendering a molecule of functional, covalently closed circular DNA (cccDNA). The HBV genome consists of four genes with four partially-overlapping ORFs: core (C), polymerase (P), surface antigen (S), and X. The core gene consists of the precore and core regions [17]. The presence of several in-frame translation initiation codons for S and C genes allows for the translation of related but functionally distinct proteins [17]. The C ORF encodes the viral nucleocapsid (HBcAg) at the core ORF and hepatitis B e antigen (HBeAg) at the precore ORF. The S ORF codes for envelope proteins, HBsAg [17]. | |||
Polymerase (pol) is a large protein and consists of three subunits: a reverse transcriptase, a terminal protein subunit involved in (-) strand synthesis, and the ribonuclease H which helps replication and breaks down pregenomic RNA [17]. | |||
Strand-specific synthesis during replication is controlled by two genomic elements, direct repeats 1 and 2 (DR1 and DR2) in the 5’ ends of the plus strand. Liver-specific expression of viral gene products is controlled by two viral enhancer elements, En1 and En2. | |||
==Replication Cycle== | |||
To enter a cell, hepatitis B virus binds an unidentified receptor and is endocytosed. Studies using duck HBV models indicate that carboxypeptidase D plays an essential role in viral entry [17]. After uncoating of the nucleocapsid core, the viral genome enters the nucleus where the single-stranded gap region is repaired by viral polymerase, leaving the viral genome as cccDNA, the functional template for transcription by host RNA polymerase II. | |||
==Hepatitis B Virus Protein X Interacts with Host and Viral Components== | |||
Since the first report of a miRNA possessing antiviral properties in 2005 [15], researchers have begun to explore the possibility of interactions between host miRNAs and HBV. These interactions have provided mechanistic insights regarding the role of HBx in host gene expression and HCC development in HBV-infected individuals. | |||
In vitro, HBx binds eukaryotic basal transcription factors essential for proper RNAP II-DNA interactions and promoter escape, including TATA-binding protein (TBP), TFIIB, TFIIH, and the RNAP subunit RPB5 [1, 11]. These interactions facilitate transcription of viral genes. HBx has been shown to suppress apoptosis of hepatocytes via induction of survivin, disruption of the tumor suppressor p53, and activation of c-jun N-terminal kinases [13, 12]. In infected cells, HBx induces overexpression of a cellular target, HBXIP, which functions as a regulator of centrosome dynamics and cytokinesis – critical aspects of mitosis [14]. Furthermore, it was reported that HBx activates src and the ras/raf/ERK (extracellular signal-regulated kinase) pathway, which leads to proliferation of quiescent cells [12]. | |||
There have been numerous studies investigating the mechanisms of HBx-mediated cell proliferation and HCC development. Two studies in the 1990s demonstrated HBx potentiates c-myc induced liver oncogenesis in a mouse model [3, 4]. HBx mediates the expression of a wide variety of host RNAs and genes, including transcription factors, the p-53 tumor-suppressor gene, oncogenes, ribosomal protein S27a [6], and ribosomal RNAs [7]. In HCC cells, overexpression of HBx and the oncogene c-myc each increased rRNA levels; when cells were co-transfected with HBx and c-myc, an additive effect on both rRNA synthesis and cell division was observed [7]. Furthermore, in vitro analyses of HCC cells infected with HBx-knockdown HBV demonstrate the existence of particular HBx-dependent proliferative signaling events in this cancer [5]. Collectively, these findings suggest that HBx is a key mediator of HCC development in HBV infected individuals. | |||
==Overview of Interactions between HBV and Host miRNAs== | ==Overview of Interactions between HBV and Host miRNAs== | ||
Several cellular transcription factors have an affinity for HBV enhancer elements and thus are crucial for efficient viral replication. A 2011 review reported C/EBP and CREB as TFs necessary for efficient viral replication. C/EBP is “CAAT enhancer-binding protein,” designated as such because of its affinity for the CAAT promoter motif, and CREB is cAMP-response element binding protein. Since publication of the review, Hu et al reported peroxisome proliferator-activated receptor alpha (PPARA) as | Several cellular transcription factors have an affinity for HBV enhancer elements and thus are crucial for efficient viral replication. A 2011 review reported C/EBP and CREB as TFs necessary for efficient viral replication. C/EBP is “CAAT enhancer-binding protein,” designated as such because of its affinity for the CAAT promoter motif, and CREB is cAMP-response element binding protein. Since publication of the review, Hu et al. reported peroxisome proliferator-activated receptor alpha (PPARA) as a host TF that binds viral enhancer elements and induces transcription of viral genes. A common method to screen for host miRNAs with HBV mRNA sequence alignment is to conduct an in silico high-throughput screen. The miRNA that targets PPARA (miRNA-141) was selected in a preliminary screening of “hit” sequences because it was the most effective inhibitor of HBV replication in a screen of 64 miRNA sequences. | ||
In an effort to elucidate the precise host-virus interactions which result in HBx-regulated miRNA expression, Ren et al. overexpressed HBc, HBs, and HBc, and HBx proteins separately in the presence of overexpressed Drosha in HCC cells. They observed decreased Drosha activity only in the presence of HBx. Inhibition of HBx by shRNA knockdown corresponded with increased Drosha activity. Considering Drosha is the key protein involved in processing functional miRNAs, this study provides strong evidence that HBx alters host gene expression through a complex network of interactions. Ren et al. provided a base of support for further study of HBx-miRNA interactions in HCC development. In 2011, Wu et al. showed that HBx down-regulates the miR-16 family in malignant hepatocytes. This family of RNAs targets various cyclins that are responsible for activating cyclin-dependent kinases, making these miRNAs key regulators of the G1/S cell-cycle checkpoint [2]. | |||
X. Wei et al. (2012) investigated HBx-mediated expression of a miRNA (miR-101) that targets DNA methyltransferase DNAMT3A [8]. The ability of HBx to down-regulate activity of a DNA methyltransferase suggests hepatitis B virus can influence epigenetic modifications to alter host gene expression. Evidence of miR-101 down-regulation in HBV-positive HCC tumor tissue suggests that miR-101 plays a tumor-suppressive role in HCC development [8]. The authors reported DNAMT3A targets several tumor-suppressor genes including RASSF1, APC, and CDKN2A. To summarize, HBx mediates increased expression of DNMT3A by decreasing miR-101 levels, thus decreasing expression of tumor-suppressor genes like RASSF1, APC, and CDKN2A. | |||
==Current HBV Therapies== | |||
The seven drugs currently approved for the treatment of chronic hepatitis B in the US and Europe include IFN-α, pegylated IFN-α, and nucleotide analogues (NUCs) lamivudine, adefovir, entecavir, telbuvudine, and tenofovir [10]. In general, interferons are capable of inhibiting viral replication and stimulating an anti-viral immune response (mobilizing NK cells activity). NUCs inhibit synthesis of the viral genome by HBV polymerase by competing with natural NTPs for polymerase uptake. The lack of a hydroxyl group on NUCs prevents polymerase from forming an additional covalent bond, effectively halting DNA synthesis. Tenofovir, an adenosine analogue, is a common component of HAART regimens used to treat HIV infected individuals. | |||
Despite extensive efforts to characterize HBx, a successful inhibitor of the viral protein has not been developed. However, according to a 2012 issue of HBV Journal Review, researchers at UC-Boulder have identified two small proteins that help inhibit cell death, Bcl-2 and Bcl-xL, as host targets of HBx [9]. The group determined that HBx interacts with these proteins to increase intracellular calcium, inducing cell proliferation and HBV DNA replication [9]. Using a C. elegans model, researchers hope to develop drugs that bind the Bcl-2/Bcl-xL recognition motif of HBx to reverse the effects of the viral oncoprotein [9]. | |||
Further research could be aimed at designing ways to restore miRNA functions (depending on the case) and especially at designing HBx inhibitors. | |||
==References== | ==References== | ||
Revision as of 22:46, 15 November 2012
A Viral Biorealm page on the family Hepatitis B Virus X gene in Hepatocellular Carcinoma
Hepatocellular carcinoma (HCC) is among the most frequently diagnosed cancers worldwide. For over forty years physicians and researchers have observed an association between HBV infection and development of HCC. Of the seven HBV genes, hepatitis b virus x protein (HBx, 17.5 kDa) is the smallest and required extensive efforts to characterize; its elusiveness is highlighted by its designation “X” [1]. The protein functions as a transcriptional transactivator of both viral and cellular transcripts, in addition inducing cytoplasmic signal transduction pathways [1]. Interestingly, HBx was a more potent inducer of transcription upon deletion of its N-terminal 1 to 50-amino-acid fragment, suggesting this domain may function as a negative regulatory element [1].
First subtopic
hep b infection, HCC
Virus Overview
Hepatitis B virus is most commonly transmitted by contact with infected blood or body fluids. The virus may also be vertically transmitted during childbirth. The infection localizes in the liver and has been linked to the development of several diseases including cirrhosis and hepatocellular carcinoma. The hepatitis B vaccine contains hepatitis B surface antigen (HBsAg). The doses of the vaccine yield a sufficient adaptive immune response capable of
Hepatitis B Virus Genome
The hepadnaviral genome is packaged as relaxed circular (RC), partially double-stranded DNA about 3.2 kb in length. After viral DNA enters the nucleus, the partially double-stranded viral DNA is repaired and the (+) DNA strand is extended, rendering a molecule of functional, covalently closed circular DNA (cccDNA). The HBV genome consists of four genes with four partially-overlapping ORFs: core (C), polymerase (P), surface antigen (S), and X. The core gene consists of the precore and core regions [17]. The presence of several in-frame translation initiation codons for S and C genes allows for the translation of related but functionally distinct proteins [17]. The C ORF encodes the viral nucleocapsid (HBcAg) at the core ORF and hepatitis B e antigen (HBeAg) at the precore ORF. The S ORF codes for envelope proteins, HBsAg [17].
Polymerase (pol) is a large protein and consists of three subunits: a reverse transcriptase, a terminal protein subunit involved in (-) strand synthesis, and the ribonuclease H which helps replication and breaks down pregenomic RNA [17].
Strand-specific synthesis during replication is controlled by two genomic elements, direct repeats 1 and 2 (DR1 and DR2) in the 5’ ends of the plus strand. Liver-specific expression of viral gene products is controlled by two viral enhancer elements, En1 and En2.
Replication Cycle
To enter a cell, hepatitis B virus binds an unidentified receptor and is endocytosed. Studies using duck HBV models indicate that carboxypeptidase D plays an essential role in viral entry [17]. After uncoating of the nucleocapsid core, the viral genome enters the nucleus where the single-stranded gap region is repaired by viral polymerase, leaving the viral genome as cccDNA, the functional template for transcription by host RNA polymerase II.
Hepatitis B Virus Protein X Interacts with Host and Viral Components
Since the first report of a miRNA possessing antiviral properties in 2005 [15], researchers have begun to explore the possibility of interactions between host miRNAs and HBV. These interactions have provided mechanistic insights regarding the role of HBx in host gene expression and HCC development in HBV-infected individuals. In vitro, HBx binds eukaryotic basal transcription factors essential for proper RNAP II-DNA interactions and promoter escape, including TATA-binding protein (TBP), TFIIB, TFIIH, and the RNAP subunit RPB5 [1, 11]. These interactions facilitate transcription of viral genes. HBx has been shown to suppress apoptosis of hepatocytes via induction of survivin, disruption of the tumor suppressor p53, and activation of c-jun N-terminal kinases [13, 12]. In infected cells, HBx induces overexpression of a cellular target, HBXIP, which functions as a regulator of centrosome dynamics and cytokinesis – critical aspects of mitosis [14]. Furthermore, it was reported that HBx activates src and the ras/raf/ERK (extracellular signal-regulated kinase) pathway, which leads to proliferation of quiescent cells [12]. There have been numerous studies investigating the mechanisms of HBx-mediated cell proliferation and HCC development. Two studies in the 1990s demonstrated HBx potentiates c-myc induced liver oncogenesis in a mouse model [3, 4]. HBx mediates the expression of a wide variety of host RNAs and genes, including transcription factors, the p-53 tumor-suppressor gene, oncogenes, ribosomal protein S27a [6], and ribosomal RNAs [7]. In HCC cells, overexpression of HBx and the oncogene c-myc each increased rRNA levels; when cells were co-transfected with HBx and c-myc, an additive effect on both rRNA synthesis and cell division was observed [7]. Furthermore, in vitro analyses of HCC cells infected with HBx-knockdown HBV demonstrate the existence of particular HBx-dependent proliferative signaling events in this cancer [5]. Collectively, these findings suggest that HBx is a key mediator of HCC development in HBV infected individuals.
Overview of Interactions between HBV and Host miRNAs
Several cellular transcription factors have an affinity for HBV enhancer elements and thus are crucial for efficient viral replication. A 2011 review reported C/EBP and CREB as TFs necessary for efficient viral replication. C/EBP is “CAAT enhancer-binding protein,” designated as such because of its affinity for the CAAT promoter motif, and CREB is cAMP-response element binding protein. Since publication of the review, Hu et al. reported peroxisome proliferator-activated receptor alpha (PPARA) as a host TF that binds viral enhancer elements and induces transcription of viral genes. A common method to screen for host miRNAs with HBV mRNA sequence alignment is to conduct an in silico high-throughput screen. The miRNA that targets PPARA (miRNA-141) was selected in a preliminary screening of “hit” sequences because it was the most effective inhibitor of HBV replication in a screen of 64 miRNA sequences. In an effort to elucidate the precise host-virus interactions which result in HBx-regulated miRNA expression, Ren et al. overexpressed HBc, HBs, and HBc, and HBx proteins separately in the presence of overexpressed Drosha in HCC cells. They observed decreased Drosha activity only in the presence of HBx. Inhibition of HBx by shRNA knockdown corresponded with increased Drosha activity. Considering Drosha is the key protein involved in processing functional miRNAs, this study provides strong evidence that HBx alters host gene expression through a complex network of interactions. Ren et al. provided a base of support for further study of HBx-miRNA interactions in HCC development. In 2011, Wu et al. showed that HBx down-regulates the miR-16 family in malignant hepatocytes. This family of RNAs targets various cyclins that are responsible for activating cyclin-dependent kinases, making these miRNAs key regulators of the G1/S cell-cycle checkpoint [2]. X. Wei et al. (2012) investigated HBx-mediated expression of a miRNA (miR-101) that targets DNA methyltransferase DNAMT3A [8]. The ability of HBx to down-regulate activity of a DNA methyltransferase suggests hepatitis B virus can influence epigenetic modifications to alter host gene expression. Evidence of miR-101 down-regulation in HBV-positive HCC tumor tissue suggests that miR-101 plays a tumor-suppressive role in HCC development [8]. The authors reported DNAMT3A targets several tumor-suppressor genes including RASSF1, APC, and CDKN2A. To summarize, HBx mediates increased expression of DNMT3A by decreasing miR-101 levels, thus decreasing expression of tumor-suppressor genes like RASSF1, APC, and CDKN2A.
Current HBV Therapies
The seven drugs currently approved for the treatment of chronic hepatitis B in the US and Europe include IFN-α, pegylated IFN-α, and nucleotide analogues (NUCs) lamivudine, adefovir, entecavir, telbuvudine, and tenofovir [10]. In general, interferons are capable of inhibiting viral replication and stimulating an anti-viral immune response (mobilizing NK cells activity). NUCs inhibit synthesis of the viral genome by HBV polymerase by competing with natural NTPs for polymerase uptake. The lack of a hydroxyl group on NUCs prevents polymerase from forming an additional covalent bond, effectively halting DNA synthesis. Tenofovir, an adenosine analogue, is a common component of HAART regimens used to treat HIV infected individuals. Despite extensive efforts to characterize HBx, a successful inhibitor of the viral protein has not been developed. However, according to a 2012 issue of HBV Journal Review, researchers at UC-Boulder have identified two small proteins that help inhibit cell death, Bcl-2 and Bcl-xL, as host targets of HBx [9]. The group determined that HBx interacts with these proteins to increase intracellular calcium, inducing cell proliferation and HBV DNA replication [9]. Using a C. elegans model, researchers hope to develop drugs that bind the Bcl-2/Bcl-xL recognition motif of HBx to reverse the effects of the viral oncoprotein [9]. Further research could be aimed at designing ways to restore miRNA functions (depending on the case) and especially at designing HBx inhibitors.
References
Beck, Juergen, and Michael Nassal. "World Journal of Gastroenterology." 13.1 (2007): 48-64.
Bouchard, MJ, and RJ Schneider. "Journal of Virology." 78.23 (2004): 12725-12734.
Marusawa, H, S Matsuzawa, et al. "The EMBO Journal."EMBO Journal. 22.11 (2003): 2729-2740.
Cheng, Alfred A.L., Jun Yu, et al. "Biochemical and Biophysical Research Communications." (2008)
Qadri, I, H Maguire, et al. "Proceedings of the National Academy of Sciences." 92. (1995): 1003-1007.
Liu, Wan-Hsin, Shiou-Hwei Yeh, et al. "Biochemica et Biophysica Acta." Biochemica et Biophysica Acta. (2011): 678-685.
Ren, Min, Dongdong Qin, et al. "Antiviral Research." 94. (2012): 225-231.
He, Yan, Hui-qing Sun, et al. "Medical Oncology." 27. (2009): 1227-1233.