Flavivirus NS1 protein: Difference between revisions
No edit summary |
|||
| (38 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
== | {{Uncurated}} | ||
[[Image: | ==Flavivirus== | ||
<br> | <br>By Alex Gonzales<br> | ||
<br> | <br>Flaviviruses are small, positive sense RNA viruses that can cause yellow fever, dengue, Japanese encephalitis, and West Nile virus in addition to tick-borne encephalitis (Hetnz and Stiasny 2012). These pathogens use arthropods such as mosquitoes from the <i>Aedes</i> genus as vectors and are found in temperate and tropical areas. The specificity of their natural hosts means that flaviviruses all have a certain endemic/ epidemic area rather than being equally distributed around the globe. However, that also means that the virus's range can expand when the host adapts to new places. For example the introduction of West Nile vector and the subsequent outbreak in New York in 1999 lead to the expansion of its target range into North and South America (Vasilakis et al. 2011). | ||
<br> | |||
Globally, the dengue virus has the most impact with about 50-100 million infections per year which result in more than 20,000 deaths annually. This is probably due to the fact that there are vaccines for yellow fever, Japanese encephalitis, and tick born encephalitis but an effective dengue vaccine has yet to be discovered. The delay in the dengue vaccine comes from concerns that the vaccines might actually predispose the subject to infections from other strains of dengue virus (Heintz, Stiasny 2012). | |||
In the search for a viable vaccine, over three decades of research has been focused on the structure and pathology of Flaviviruses. Over the course of their work, scientists have discovered much, but many questions still remain about an enigmatic nonstructural glycoprotein, NS1. NS1 was discovered in 1970 in the serum of a patient infected with dengue and it was later discovered that all flavivirus NS1 genes were highly homologous. Additionally, it was found that NS1 has multiple oligomeric forms, a dimer and a hexamer, which are involved in replication and host immune system evasion respectively. It has also been suggested that the two main forms of NS1 are involved in both protection and pathogenesis of their host (Muller, Young 2013). <br> | |||
<br> | |||
<br> | ==Basic Flavivirus Structure and Maturation== | ||
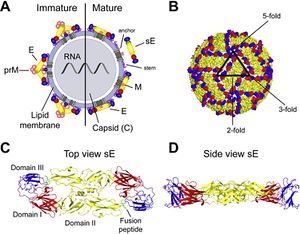
<br> | [[Image:Alex Fig 1.jpeg|thumb|300px|right|Figure 1. Flavivirus Structure. (A) Schematic representation of a flavivirus particle. Left: immature virion; right: mature virion.(B) Herringbone-like arrangement of 90 E protein dimers at the virion surface as determined by cryo-electron microscopy.(Heinz and Stiasny 2012]] | ||
<br><br> | |||
<br> Flaviviruses are small and have only seven nonstructural and three structural proteins. These structural proteins are designated E, prM/M, and C for envelope, precursor of the membrane or membrane, and capsid respectively (Fig 1). Immature virions are formed by budding of the endoplasmic reticulum. These non-infections virions form prM-E heterodimers. In order to mature into infectious virions, these heterodimers need to be cleaved. This allows the E proteins to form herringbone like antiparallel dimers and therefore rearranges the whole viral envelope into a smoother surface (Heinz and Stiasny 2012). | |||
E proteins are important for cell infection as they mediate binding to cell receptors and fusion with endosomal membranes after host cell endocytosis. To help fusion with the endosomal membrane, E proteins undergo yet another structural change triggered by pH. In low pH, the E protein dimer disassociates, and becomes a trimer with an exposed fusion peptide at one tip. Due to the essential role E proteins play in entering the host cell, they are a main target of anti-flivivrus antibodies which attempt to block entry into the cell by hindering attachment and endocytosis or membrane fusion (Heinz and Stiasny 2012). | |||
==NS1 Structure== | |||
[[Image:Alex Fig 2.jpeg|thumb|300px|left|Figure 2. NS1 dimer structure. (A) NS1 dimer with one subunit in gray and the other colored by domain (blue, b roll; yellow, wing with orange connector subdomain; red, central b ladder). Disulfides are shown as yellow spheresand N-linked glycosylation sites as black sticks. A 20-residue disordered regions indicated with dotted lines. C, C terminus; N, N terminus.(Akey et al 2014). ]] | |||
<br>All flavivirus NS1 genes encode a 352 amino acid polypeptide with a slight variation in glycosylation status which gives it a variable weight of 46-55 kDa (Muller and Young 2013). Up until recently, the structure of NS1 was unknown due to its dissimilarity with known sequences and an inability to produce pure, stable proteins. Akey et al.(2014) finally elucidated the structure for the two main forms of NS1 using a baclovirus expression system in insect cells which produced full length, recombinant, glycosylated West Nile and dengue type two virus NS1. In their NS1 dimer structure, all amino acids are accounted for except a twenty amino acid stretch from amino acid 108-128 (Akey et al 2014). | |||
Shown in Figure 2, the NS1 dimer contains a central β-sheet domain with two other domains. The first is a small β-roll domain that is a domain swap structure of two β hairpins that are stabilized with a disulfide bond. These hairpins reach across the axis and intertwine, forming a curved β-sheet as indicated in blue in figure 2A. The monomer’s second domain resembles a wing coming off of the central β-sheet. The wing, highlighted in yellow (3A), has two glycosylation sites, an internal disulfide, and contains two discreet domains. One of these domains is an α/ β subdomain which is made up of a four stranded β-sheet, two α helixes, and a disordered distal tip which is represented in Fig 2A as a dotted line. The other subdomain is a discontinuous connector that sits against the β-roll and is shown in orange. NS1’s third domain is the most noticeable as it is a continuous β-sheet that runs along the whole length of the dimer and resembles a ladder. It has 18 antiparallel β strands that look like rungs with each monomer contributing exactly nine rungs. There also happens to be a long “spaghetti loop” between β13 and β14 that holds it shape through hydrogen bonds. Overall, the dimer 90 A long, has a 90A “wingspan” and is 40A in the third dimension (Akey et al. 2014). | |||
Thanks to a protrusion created by the β-roll and connector subdomain interaction, one side of the NS1 dimer is hydrophobic. Additionally, a dipeptide that has been suggested to interact with the NS4B transmembrane protein is located along the edge of the protrusion. This plus the hydrophobicity could allow the NS1 dimer to interact with the ER membrane and NS4A and B, which are a part of the replication complex along with the other six nonstructural proteins (Atkey et al. 2014). | |||
NS1 dimers are thought to be intracellular, while the other form, the hexamer, is thought to be secreted. The hexameric NS1 is a soluble, proteolipid particle that seems to have configurations that vary between and among the West Nile and dengue viruses. Akey et al. (2014) found three different hexamer configurations but they noted that all hexamers were made up of three dimers. They also followed a trend where the three β rolls were facing the interior with the spaghetti loops, glycosylation sites, and wing domain loops were facing outward. <br> | |||
==NS1 Expression, post translational processing and trafficking == | |||
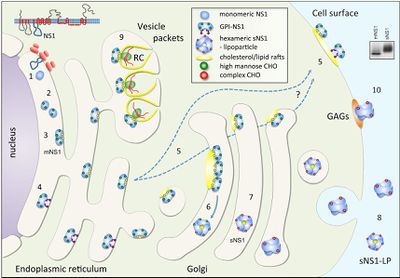
[[Image:Alex Fig 3.jpeg|thumb|400px|right|Figure 3.Schematic summary of NS1 trafficking in mammalian cells. (Muller and Young 2013) ]] | |||
<br>NS1 is translated during the first round of translation and is translocated to the ER lumen for processing (Falgout et al.1989). In the ER, it is cleaved in various places by an unknown host signal proteins and glycosylated by high-mannose carbohydrates. After that, it promptly dimerizes (20-40min). This is most likely because of the slightly hydrophobic elements mentioned above (Winkler et al. 1988). The newly dimerized NS1 has one of three destinations: a viral replication site somewhere in the cell, the cell surface, or it is secreted into the extracellular space (Figure 3). The NS1 that stays in the cell generally co-localizes with dsRNA and the other nonstructural proteins which are involved in genome replication within structures called vesicle packets. A much smaller percent of the dimer tends to localize to the surface of the infected cell. The hexamer form of NS1 is only found in mammalian cells and comes from NS1 sent to the Golgi for processing (Muller and Young 2013). In the Golgi, complex carbohydrates and sugars are added and some trimming of mannose occurs. | |||
The NS1 hexamer isn't found in infected mosquitoes because insects don’t have the machinery required for this process (Muller and Young 2013). However there is one report, Ludert et al. (2008) that claims the NS1 is secreted from insect cells. Yet this report may be slightly flawed because it is based on NS1 detection in media with a NS1 ELISA capture assay and the results were qualitative rather than quantitative. Additionally, the results were gathered late in infection so there were probably some cells that lysed which would secrete detectable amounts of NS1 into the media (Ludert et al. 2008). In insect cells, NS1 just accumulates which may suggest a link between secretion and the addition of carbohydrates (Muller and Young 2013). | |||
Despite the NS1 protein’s genetic homology between species, interspecies retention and secretion rates tend to differ. Using immunofluorescence, Youn et al. (2010) showed that West Nile Virus NS1 accumulated at higher levels on the cell surface relative to dengue NS1 and that dengue NS1 had higher levels of secretion. Youn et al. (2010) suggested that this difference in secretion was due a motif at the N terminus of the protein that may affect differential binding to some ER resident host cell protein which would affect the final destination of the NS1 protein. However, it might also be possible that the motif could affect hexamer formation making the dengue NS1 more efficient at hexamer formation. This would make the higher rate of membrane binding a loss of function mutation. | |||
Detailed compositional analysis of the NS1 hexamer revealed that it carries equal amounts of diacylglycerol, cholesterol, choresteryl ester, phosphatidalychlorine , phosphatidyl-ethanolamine and sphingomyelin (Gutsche et al. 2011). Gutsche et al (2011), suggests that the tight association of these lipids could lead to a mechanism for the hexamer formation. They believe that once the dimeric NS1 protein associates with the membrane the dimers cluster together at lipid rafts, lipid droplets, and or triglycerides and this disrupts the membrane with three dimers pinching off to form a hexamer (Gustsche et al 2011). The pinching method would account for the lipid content of the hexamers. There is actually support for this model as it has been reported that treating cells with lipid-modulating drugs decreases NS1 hexamer secretion (Gustsche et al 2011). <br> | |||
==NS1 as a cofactor in virus replication == | |||
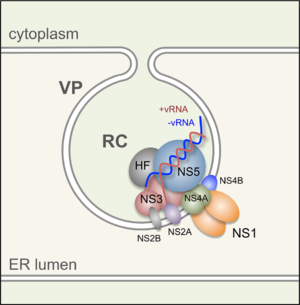
[[Image:Alex_Fig_4.png|thumb|300px|left|Figure 4. Flavivirus replication complex(Muller and Young 2013) ]] | |||
<br> The flavivirus polyprotein has specific transmembrane domains that determine whether the individual proteins are going to be located on the cytoplasmic or luminal side of the endoplasmic reticulum (ER) after cleavage of the polyprotein . After cleavage, C, NS3 and NS5 are on the cytoplasmic side while PrM, E, and NS1 end up in the lumen. With the exception of short regions between transmembrane domains, the NS2A, NS2B, NS4A and NS4B are stuck in between the lipid bilayer of the ER (Brinton 2014). | |||
The exact function of NS2A, NS2B, NS4A, and NS4B is not known as their conserved domains are dissimilar to those of known enzymes. Little is known about the structure of these proteins. However, researchers have discovered that they are small, hydrophobic proteins with two or three membrane spanning regions that facilitate the assembly and or anchoring of the viral replication complexes to the ER membrane. It is believed that they interact with one another to form the viral replication complex (Figure 5). The C terminus of NS3 has been found to interact with the N terminus of NS5. NS5 only interacts with NS3, but NS3 also interacts with a membrane associated co-factor NS2B. NS2B is integral to the viral replication complex as it interacts with three other membrane associated proteins: NS2A, NS4A and NS4B. Additionally, a NS42A – NS2A interaction has been observed, but neither a NS2A-NS4B nor a NS4A-NS4B interaction has been observed (Brinton 2014). However, NS4B and NS4A have been shown to interact on some level with NS1 (Atkey et al 2014)(Makenzie et al 1996). | |||
At first, due to its similar secretion profile to E and prM, it was thought that NS1 was involved in the virion assembly and maturation process (Muller and Young 2013). However, that theory was eventually proven wrong and as it was shown that NS1 localized to viral RNA replication sites in invaginations of the ER (Mackenzie et al. 1996). This suggests a role in RNA replication, possibly as a member of the viral replication complex but NS1 localizes to the luminal side of the ER, the opposite side for all of the other replication machinery (Muller and Young 2013) (Figure 5). To determine its role in viral replication, various mutations have been made to the NS1 protein. For example, a mutation in the NS1-N-linked glycosylation sites in the yellow fever genome dramatically reduced viral RNA replication levels. A similar experiment created a large frame mutation in the yellow fever genome NS1 gene which resulted in the inability to replicate viral RNA. The ability was rescued by the addition of NS1 in trans which suggests that NS1 is required for the initiation of RNA synthesis and is possibly an essential part of early minus strand synthesis (Brinton 2014). | |||
Minus strand synthesis happens early in the viral life cycle when there are a low number of genomes. These genomes switch back and forth between functioning as an mRNA used in translation and a template for minus strand RNA synthesis. Eventually, the minus strand templates are sequestered in the ER membrane invaginations and the associate with replication complexes (Brinton 2014). Being in the lumen of the ER, and with its possible association with the replication complex, it is quite plausible that NS1 is a part of minus strand synthesis. | |||
It has been suggested that due to its luminal localization, NS1 plays a more structural support role. Just looking at the structure of the NS1 dimer, it was suggested that it was able to interact with NS4A and NS4B (Atkey et al 2014). It has been found that the NS1-NS4B interaction is necessary for early viral RNA synthesis (Brinton 2014). Makenzie et al (1996) found that NS1 physically associated with the column bound NS4A. Additionally, through substitution experiments, Youn et al. (2012) were able to show that there is a physical and genetic nteraction between NS1 and NS4B as well. However, they were unsure as to whether this interaction was a direct or an indirect one They concede the probability that the other proteins in the complex may mediate the physical interaction between NS1 and NS4B (Youn et al. 2012). Looking at these combined results, Muller and Young (2013) suggest that from the lumen of the ER, NS1physically interacts with both NS4 proteins providing stability for the replication complex making its role more of a structural one. | |||
<br> | |||
==NS1 engagement with immune system == | |||
<br> The NS1 secreted hexamer is highly circulated during the acute stage of infection, so it isn’t surprising that it is an immunogen. For example, in a primary dengue infection there are low anit-NS1 IgM and IgG responses but in a secondary infection there is a rapid rise in anamestic antibody repose early on (Muller and Young 2013). These antibodies aren’t neutralizing because NS1 isn’t a virion component. However, in the 1980s, it was found that flavivirus specific monocolonal antibodies (MAbs) could actually protect mice from dying of yellow fever in passive protection studies. Further experimentation suggested that the antibodies recognized the surface NS1 and initiated complement mediated lysis of infected cells. More recently, studies have shown that protection in mice could also be a result of phagocytosis and receptor recognition and have given scientists a better understanding of anti-NS1 antibodies and their role in providing protection against flaviviruses (Muller and Young 2013). | |||
Recently, there has been a renewed interest in using NS1 as an element of flavivirus vaccines. In the past, there have been attempts to deliver NS1 as a vaccine candidate but they were unsuccessful. Scientists have tried immunization with recombinant proteins, live recombinant vaccine virus, recombinant baculovirus derived, naked DNA, and peptide dependent and independent anit-body protection (Muller and Young 2013). However, there are pretty significant problems with using NS1 in a vaccine. One such problem is that while the dengue NS1 protein does elicit protection, it also does a lot of damage. Some epitopes cause the immune system to create auto-antibodies that then cross react to platelets and components of the extra cellular matrix in addition to binding and inducing damage to epithelial cells (Muller and Young 2013). In essence, sometimes these vaccines end up enhancing the disease. | |||
The NS1 activated auto-antibodies seem to be responsible for a lot of the damage that is characteristic of flavivirus diseases. An early induction of these auto-antibodies in secondary dengue infection causes massive damage to uninfected epithelial cells and suggests a role for these anti-bodies in the endothelial cell dysfunction that causes vascular leaking and hemorrhaging in patients with more extreme cases of dengue (Muller and Young 2013). Additionally it was found that in mice, that these auto antibodies bound to liver epithelium and lead to cell damage and monocyte infiltration. Liver damage is characteristic of dengue virus infections and the damage done to the mouse livers supports the hypothesis that these antibodies play a key role in causing the liver damage seen in human patients as well (Muller and Young 2013). | |||
Yet there are cases where patients make a dramatic recovery from symptoms such as vascular leaking and it does not coincide with a drop in auto antibody levels. Furthermore, these raised auto-antibody levels are only found in secondary infections, as babies suffering from dengue do not have high levels of auto antibodies in circulation during the acute phase of the disease (Muller and Young 2013). This goes to show that while these highly reactive antibodies might play some role in flavivirus pathogenesis, other factors need to be taken into consideration before any real conclusions can be drawn. <br> | |||
==NS1 as a diagnostic marker== | |||
[[Image:ELSIA kit.jpg|thumb|300px|right|Figure 5. NS1 ELISA kit (http://rapidtest.com/blog/tag/dengue-ns1-elisa-kit/) ]] | |||
<br> As flaviviruses tend to co-circulate in certain areas, it can be difficult to determine which serotype patients are infected with (Heinz and Stiansy 2012). Generally, IgM ELISAs are used but they have trouble discerning different serotypes, and they aren’t effective early in the infection. NS1 however, is secreted early in infections and in large enough amounts to be detected by ELSA. Assessment of an NS1 assay from various groups led to the development of multiple NS1 capture ELISAs (Fig 5). These assays have been through multiple field tests and have been deemed valuable in the early detection of a dengue infection. NS1 can be detected in serum or plasma samples as early as 9-12 days after disease onset. Unfortunately, in secondary dengue infections, antibodies clear NS1 from circulation in 5-7 days so it is fairly likely the test would result in a false negative. This makes it less practical in endemic countries where most of the patients would have secondary infections (Muller and Young 2013). <br> | |||
==References== | |||
[http://www.cdc.gov/vhf/virus-families/flaviviridae.html Akey DL, Brown W.C, Dutta S., Konwerski J. , Jose J., Jurkiw T.J, DelProposto J, Ogata C.M,Skiniotis G., Kuhn R.J, J.L. Smith. 2014. Flavivirus NS1 Structures Reveal Surfaces for Associations with Membranes and the Immune System. Science vol343: 881-885.] | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3917430/ Brinton MA.2014. Replication cycle and molecular biology of the West Nile Virus.Viruses 6(1): 13-53.] | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC250595/ Falgout B, Chanock R, CJ Lai. 1989. Proper processing of dengue virus nonstructural glycoprotein NS1 requires the N-terminal hydrophobic signal sequence and the downstream nonstructural protein NS2A. J Virol 63(5): 1852-1860.] | |||
[http://www.sciencedirect.com/science/article/pii/S1386653212003435 F.X. Heinz, K. Stiansy.2012. Flaviviruses and their antigenic structure. Journal of Clinical Virology 55: 289-295.] | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3093454/ Gutsche I, Coulibaly F, Voss JE, Salmon J, d’Alayer J, Ermonval M, Larquet E, Charnue P, Krey T, Merget F, Guittet E, Rey FA, Flamand M. 2011. Secreted dengue virus nonstructural protein NS1 is an atypical barrel-shaped high-density lipoprotein. PNAS 108(19): 8003-8008.] | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2386455/ Ludert JE, Mosso C, Ceballos-Olvera I, del Angel RM. 2008. Virol J 5: 51] | |||
[http://www.sciencedirect.com/science/article/pii/S0042682296903074 Mackenzie JM, Jones MK, Young PR. 1996. Immunolocalization of the dengue virus nonstructural Gylcoprotein NS1 suggests a role in Viral RNA replication. Virology 220(1): 232-240.] | |||
[http://www.sciencedirect.com/science/article/pii/S0166354213000624 Muller D.A., P.R Young, 2013. The Flavivirus NS1 protein: Molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antiviral Research 98: 192-208.] | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3321645/pdf/nihms-368054.pdf Vasilakis N., Cardosa J., Hanly K.A, HolmesE.C., S.C Weave , (2011) Fever from the forest: prospects for the continued emergence of sylvatic dengue virus and its impact on public health. Nat Rev Mocrobiol 9(7): 532-541.] | |||
[http://www.sciencedirect.com/science/article/pii/0042682288904084 Winkler G, Randolph VB, Cleaves GR, Ryan TE, StollarV.1988. Evidence that the mature form of the flavivirus nonstructural protein NS1 is a dimer. Virology 162(1): 187-96.] | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2937659/ Youn S, Cho H, Fremont D, Diamond MS. 2010. A short terminal peptide motif on flavivirus nonstructural protein NS1 modulates cellular targeting and immune recognition. J Virol 84(18): 9516-9532.] | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3416313/ YounS, Li T, McCune BT, Edeling MA, Fremont DH, Cristea IM, Diamond MS. 2012. Evidence for a genetic and physical interaction between nonstructural proteins NS1 and NS4B that modulates replication of West Nile virus. J Virol 86(13):7360-71.] | |||
[ | |||
Latest revision as of 13:28, 1 October 2015
Flavivirus
By Alex Gonzales
Flaviviruses are small, positive sense RNA viruses that can cause yellow fever, dengue, Japanese encephalitis, and West Nile virus in addition to tick-borne encephalitis (Hetnz and Stiasny 2012). These pathogens use arthropods such as mosquitoes from the Aedes genus as vectors and are found in temperate and tropical areas. The specificity of their natural hosts means that flaviviruses all have a certain endemic/ epidemic area rather than being equally distributed around the globe. However, that also means that the virus's range can expand when the host adapts to new places. For example the introduction of West Nile vector and the subsequent outbreak in New York in 1999 lead to the expansion of its target range into North and South America (Vasilakis et al. 2011).
Globally, the dengue virus has the most impact with about 50-100 million infections per year which result in more than 20,000 deaths annually. This is probably due to the fact that there are vaccines for yellow fever, Japanese encephalitis, and tick born encephalitis but an effective dengue vaccine has yet to be discovered. The delay in the dengue vaccine comes from concerns that the vaccines might actually predispose the subject to infections from other strains of dengue virus (Heintz, Stiasny 2012).
In the search for a viable vaccine, over three decades of research has been focused on the structure and pathology of Flaviviruses. Over the course of their work, scientists have discovered much, but many questions still remain about an enigmatic nonstructural glycoprotein, NS1. NS1 was discovered in 1970 in the serum of a patient infected with dengue and it was later discovered that all flavivirus NS1 genes were highly homologous. Additionally, it was found that NS1 has multiple oligomeric forms, a dimer and a hexamer, which are involved in replication and host immune system evasion respectively. It has also been suggested that the two main forms of NS1 are involved in both protection and pathogenesis of their host (Muller, Young 2013).
Basic Flavivirus Structure and Maturation
Flaviviruses are small and have only seven nonstructural and three structural proteins. These structural proteins are designated E, prM/M, and C for envelope, precursor of the membrane or membrane, and capsid respectively (Fig 1). Immature virions are formed by budding of the endoplasmic reticulum. These non-infections virions form prM-E heterodimers. In order to mature into infectious virions, these heterodimers need to be cleaved. This allows the E proteins to form herringbone like antiparallel dimers and therefore rearranges the whole viral envelope into a smoother surface (Heinz and Stiasny 2012).
E proteins are important for cell infection as they mediate binding to cell receptors and fusion with endosomal membranes after host cell endocytosis. To help fusion with the endosomal membrane, E proteins undergo yet another structural change triggered by pH. In low pH, the E protein dimer disassociates, and becomes a trimer with an exposed fusion peptide at one tip. Due to the essential role E proteins play in entering the host cell, they are a main target of anti-flivivrus antibodies which attempt to block entry into the cell by hindering attachment and endocytosis or membrane fusion (Heinz and Stiasny 2012).
NS1 Structure
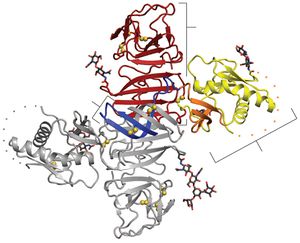
All flavivirus NS1 genes encode a 352 amino acid polypeptide with a slight variation in glycosylation status which gives it a variable weight of 46-55 kDa (Muller and Young 2013). Up until recently, the structure of NS1 was unknown due to its dissimilarity with known sequences and an inability to produce pure, stable proteins. Akey et al.(2014) finally elucidated the structure for the two main forms of NS1 using a baclovirus expression system in insect cells which produced full length, recombinant, glycosylated West Nile and dengue type two virus NS1. In their NS1 dimer structure, all amino acids are accounted for except a twenty amino acid stretch from amino acid 108-128 (Akey et al 2014).
Shown in Figure 2, the NS1 dimer contains a central β-sheet domain with two other domains. The first is a small β-roll domain that is a domain swap structure of two β hairpins that are stabilized with a disulfide bond. These hairpins reach across the axis and intertwine, forming a curved β-sheet as indicated in blue in figure 2A. The monomer’s second domain resembles a wing coming off of the central β-sheet. The wing, highlighted in yellow (3A), has two glycosylation sites, an internal disulfide, and contains two discreet domains. One of these domains is an α/ β subdomain which is made up of a four stranded β-sheet, two α helixes, and a disordered distal tip which is represented in Fig 2A as a dotted line. The other subdomain is a discontinuous connector that sits against the β-roll and is shown in orange. NS1’s third domain is the most noticeable as it is a continuous β-sheet that runs along the whole length of the dimer and resembles a ladder. It has 18 antiparallel β strands that look like rungs with each monomer contributing exactly nine rungs. There also happens to be a long “spaghetti loop” between β13 and β14 that holds it shape through hydrogen bonds. Overall, the dimer 90 A long, has a 90A “wingspan” and is 40A in the third dimension (Akey et al. 2014).
Thanks to a protrusion created by the β-roll and connector subdomain interaction, one side of the NS1 dimer is hydrophobic. Additionally, a dipeptide that has been suggested to interact with the NS4B transmembrane protein is located along the edge of the protrusion. This plus the hydrophobicity could allow the NS1 dimer to interact with the ER membrane and NS4A and B, which are a part of the replication complex along with the other six nonstructural proteins (Atkey et al. 2014).
NS1 dimers are thought to be intracellular, while the other form, the hexamer, is thought to be secreted. The hexameric NS1 is a soluble, proteolipid particle that seems to have configurations that vary between and among the West Nile and dengue viruses. Akey et al. (2014) found three different hexamer configurations but they noted that all hexamers were made up of three dimers. They also followed a trend where the three β rolls were facing the interior with the spaghetti loops, glycosylation sites, and wing domain loops were facing outward.
NS1 Expression, post translational processing and trafficking
NS1 is translated during the first round of translation and is translocated to the ER lumen for processing (Falgout et al.1989). In the ER, it is cleaved in various places by an unknown host signal proteins and glycosylated by high-mannose carbohydrates. After that, it promptly dimerizes (20-40min). This is most likely because of the slightly hydrophobic elements mentioned above (Winkler et al. 1988). The newly dimerized NS1 has one of three destinations: a viral replication site somewhere in the cell, the cell surface, or it is secreted into the extracellular space (Figure 3). The NS1 that stays in the cell generally co-localizes with dsRNA and the other nonstructural proteins which are involved in genome replication within structures called vesicle packets. A much smaller percent of the dimer tends to localize to the surface of the infected cell. The hexamer form of NS1 is only found in mammalian cells and comes from NS1 sent to the Golgi for processing (Muller and Young 2013). In the Golgi, complex carbohydrates and sugars are added and some trimming of mannose occurs.
The NS1 hexamer isn't found in infected mosquitoes because insects don’t have the machinery required for this process (Muller and Young 2013). However there is one report, Ludert et al. (2008) that claims the NS1 is secreted from insect cells. Yet this report may be slightly flawed because it is based on NS1 detection in media with a NS1 ELISA capture assay and the results were qualitative rather than quantitative. Additionally, the results were gathered late in infection so there were probably some cells that lysed which would secrete detectable amounts of NS1 into the media (Ludert et al. 2008). In insect cells, NS1 just accumulates which may suggest a link between secretion and the addition of carbohydrates (Muller and Young 2013).
Despite the NS1 protein’s genetic homology between species, interspecies retention and secretion rates tend to differ. Using immunofluorescence, Youn et al. (2010) showed that West Nile Virus NS1 accumulated at higher levels on the cell surface relative to dengue NS1 and that dengue NS1 had higher levels of secretion. Youn et al. (2010) suggested that this difference in secretion was due a motif at the N terminus of the protein that may affect differential binding to some ER resident host cell protein which would affect the final destination of the NS1 protein. However, it might also be possible that the motif could affect hexamer formation making the dengue NS1 more efficient at hexamer formation. This would make the higher rate of membrane binding a loss of function mutation.
Detailed compositional analysis of the NS1 hexamer revealed that it carries equal amounts of diacylglycerol, cholesterol, choresteryl ester, phosphatidalychlorine , phosphatidyl-ethanolamine and sphingomyelin (Gutsche et al. 2011). Gutsche et al (2011), suggests that the tight association of these lipids could lead to a mechanism for the hexamer formation. They believe that once the dimeric NS1 protein associates with the membrane the dimers cluster together at lipid rafts, lipid droplets, and or triglycerides and this disrupts the membrane with three dimers pinching off to form a hexamer (Gustsche et al 2011). The pinching method would account for the lipid content of the hexamers. There is actually support for this model as it has been reported that treating cells with lipid-modulating drugs decreases NS1 hexamer secretion (Gustsche et al 2011).
NS1 as a cofactor in virus replication
The flavivirus polyprotein has specific transmembrane domains that determine whether the individual proteins are going to be located on the cytoplasmic or luminal side of the endoplasmic reticulum (ER) after cleavage of the polyprotein . After cleavage, C, NS3 and NS5 are on the cytoplasmic side while PrM, E, and NS1 end up in the lumen. With the exception of short regions between transmembrane domains, the NS2A, NS2B, NS4A and NS4B are stuck in between the lipid bilayer of the ER (Brinton 2014).
The exact function of NS2A, NS2B, NS4A, and NS4B is not known as their conserved domains are dissimilar to those of known enzymes. Little is known about the structure of these proteins. However, researchers have discovered that they are small, hydrophobic proteins with two or three membrane spanning regions that facilitate the assembly and or anchoring of the viral replication complexes to the ER membrane. It is believed that they interact with one another to form the viral replication complex (Figure 5). The C terminus of NS3 has been found to interact with the N terminus of NS5. NS5 only interacts with NS3, but NS3 also interacts with a membrane associated co-factor NS2B. NS2B is integral to the viral replication complex as it interacts with three other membrane associated proteins: NS2A, NS4A and NS4B. Additionally, a NS42A – NS2A interaction has been observed, but neither a NS2A-NS4B nor a NS4A-NS4B interaction has been observed (Brinton 2014). However, NS4B and NS4A have been shown to interact on some level with NS1 (Atkey et al 2014)(Makenzie et al 1996).
At first, due to its similar secretion profile to E and prM, it was thought that NS1 was involved in the virion assembly and maturation process (Muller and Young 2013). However, that theory was eventually proven wrong and as it was shown that NS1 localized to viral RNA replication sites in invaginations of the ER (Mackenzie et al. 1996). This suggests a role in RNA replication, possibly as a member of the viral replication complex but NS1 localizes to the luminal side of the ER, the opposite side for all of the other replication machinery (Muller and Young 2013) (Figure 5). To determine its role in viral replication, various mutations have been made to the NS1 protein. For example, a mutation in the NS1-N-linked glycosylation sites in the yellow fever genome dramatically reduced viral RNA replication levels. A similar experiment created a large frame mutation in the yellow fever genome NS1 gene which resulted in the inability to replicate viral RNA. The ability was rescued by the addition of NS1 in trans which suggests that NS1 is required for the initiation of RNA synthesis and is possibly an essential part of early minus strand synthesis (Brinton 2014).
Minus strand synthesis happens early in the viral life cycle when there are a low number of genomes. These genomes switch back and forth between functioning as an mRNA used in translation and a template for minus strand RNA synthesis. Eventually, the minus strand templates are sequestered in the ER membrane invaginations and the associate with replication complexes (Brinton 2014). Being in the lumen of the ER, and with its possible association with the replication complex, it is quite plausible that NS1 is a part of minus strand synthesis.
It has been suggested that due to its luminal localization, NS1 plays a more structural support role. Just looking at the structure of the NS1 dimer, it was suggested that it was able to interact with NS4A and NS4B (Atkey et al 2014). It has been found that the NS1-NS4B interaction is necessary for early viral RNA synthesis (Brinton 2014). Makenzie et al (1996) found that NS1 physically associated with the column bound NS4A. Additionally, through substitution experiments, Youn et al. (2012) were able to show that there is a physical and genetic nteraction between NS1 and NS4B as well. However, they were unsure as to whether this interaction was a direct or an indirect one They concede the probability that the other proteins in the complex may mediate the physical interaction between NS1 and NS4B (Youn et al. 2012). Looking at these combined results, Muller and Young (2013) suggest that from the lumen of the ER, NS1physically interacts with both NS4 proteins providing stability for the replication complex making its role more of a structural one.
NS1 engagement with immune system
The NS1 secreted hexamer is highly circulated during the acute stage of infection, so it isn’t surprising that it is an immunogen. For example, in a primary dengue infection there are low anit-NS1 IgM and IgG responses but in a secondary infection there is a rapid rise in anamestic antibody repose early on (Muller and Young 2013). These antibodies aren’t neutralizing because NS1 isn’t a virion component. However, in the 1980s, it was found that flavivirus specific monocolonal antibodies (MAbs) could actually protect mice from dying of yellow fever in passive protection studies. Further experimentation suggested that the antibodies recognized the surface NS1 and initiated complement mediated lysis of infected cells. More recently, studies have shown that protection in mice could also be a result of phagocytosis and receptor recognition and have given scientists a better understanding of anti-NS1 antibodies and their role in providing protection against flaviviruses (Muller and Young 2013).
Recently, there has been a renewed interest in using NS1 as an element of flavivirus vaccines. In the past, there have been attempts to deliver NS1 as a vaccine candidate but they were unsuccessful. Scientists have tried immunization with recombinant proteins, live recombinant vaccine virus, recombinant baculovirus derived, naked DNA, and peptide dependent and independent anit-body protection (Muller and Young 2013). However, there are pretty significant problems with using NS1 in a vaccine. One such problem is that while the dengue NS1 protein does elicit protection, it also does a lot of damage. Some epitopes cause the immune system to create auto-antibodies that then cross react to platelets and components of the extra cellular matrix in addition to binding and inducing damage to epithelial cells (Muller and Young 2013). In essence, sometimes these vaccines end up enhancing the disease.
The NS1 activated auto-antibodies seem to be responsible for a lot of the damage that is characteristic of flavivirus diseases. An early induction of these auto-antibodies in secondary dengue infection causes massive damage to uninfected epithelial cells and suggests a role for these anti-bodies in the endothelial cell dysfunction that causes vascular leaking and hemorrhaging in patients with more extreme cases of dengue (Muller and Young 2013). Additionally it was found that in mice, that these auto antibodies bound to liver epithelium and lead to cell damage and monocyte infiltration. Liver damage is characteristic of dengue virus infections and the damage done to the mouse livers supports the hypothesis that these antibodies play a key role in causing the liver damage seen in human patients as well (Muller and Young 2013).
Yet there are cases where patients make a dramatic recovery from symptoms such as vascular leaking and it does not coincide with a drop in auto antibody levels. Furthermore, these raised auto-antibody levels are only found in secondary infections, as babies suffering from dengue do not have high levels of auto antibodies in circulation during the acute phase of the disease (Muller and Young 2013). This goes to show that while these highly reactive antibodies might play some role in flavivirus pathogenesis, other factors need to be taken into consideration before any real conclusions can be drawn.
NS1 as a diagnostic marker

As flaviviruses tend to co-circulate in certain areas, it can be difficult to determine which serotype patients are infected with (Heinz and Stiansy 2012). Generally, IgM ELISAs are used but they have trouble discerning different serotypes, and they aren’t effective early in the infection. NS1 however, is secreted early in infections and in large enough amounts to be detected by ELSA. Assessment of an NS1 assay from various groups led to the development of multiple NS1 capture ELISAs (Fig 5). These assays have been through multiple field tests and have been deemed valuable in the early detection of a dengue infection. NS1 can be detected in serum or plasma samples as early as 9-12 days after disease onset. Unfortunately, in secondary dengue infections, antibodies clear NS1 from circulation in 5-7 days so it is fairly likely the test would result in a false negative. This makes it less practical in endemic countries where most of the patients would have secondary infections (Muller and Young 2013).
References
Brinton MA.2014. Replication cycle and molecular biology of the West Nile Virus.Viruses 6(1): 13-53.
Ludert JE, Mosso C, Ceballos-Olvera I, del Angel RM. 2008. Virol J 5: 51