Eubacterium Nodatum: Difference between revisions
No edit summary |
No edit summary |
||
| (15 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | {{Uncurated}} | ||
= | =Classification= | ||
==a. Higher order taxa== | ==a. Higher order taxa== | ||
Bacteria, Firmicutes, Clostridia, Clostridiale, Clostridiales Family XIII, Eubacterium | Bacteria, Firmicutes, Clostridia, Clostridiale, Clostridiales Family XIII, Eubacterium [[#References |[1]]] | ||
= | |||
[[File:Figure 1.png|200px|thumb|left|Figure 1. Animated visual of genus Eubacterium cell structure. | |||
Source: https://en.wikipedia.org/wiki/Bacteria]] | |||
= Description and significance= | |||
Eubacterium nodatum is one of the predominant bacteria found in peri-implant sites [[#References |[2]]]. E. nodatum is a species of Eubacterium [Figure 1] that has commonly been found in patients with chronic periodontal disease. E. nodatum is obligately anaerobic, and a Gram-positive microbe [[#References |[3]]]. Although this bacterium was initially identified in the late 1980s, it has been difficult to understand its metabolic pathways since it is challenging to isolate, has a slow growth process, and is structurally complex [[#References |[4]]]. [[#References |[5]]]. According to the Centers for Disease Control and Prevention (CDC), 47.2% of American adults suffer from periodontitis, a bacterial infection that forms on oral surfaces and damages the tissues that surround teeth in peri-implant patients [[#References |[6]]]. Because E. nodatum is a periodontal pathogen, research has been focused on learning about oral microbiota, and how changes in these microbiota affect and are affected by different diseases. | Eubacterium nodatum is one of the predominant bacteria found in peri-implant sites [[#References |[2]]]. E. nodatum is a species of Eubacterium [Figure 1] that has commonly been found in patients with chronic periodontal disease. E. nodatum is obligately anaerobic, and a Gram-positive microbe [[#References |[3]]]. Although this bacterium was initially identified in the late 1980s, it has been difficult to understand its metabolic pathways since it is challenging to isolate, has a slow growth process, and is structurally complex [[#References |[4]]]. [[#References |[5]]]. According to the Centers for Disease Control and Prevention (CDC), 47.2% of American adults suffer from periodontitis, a bacterial infection that forms on oral surfaces and damages the tissues that surround teeth in peri-implant patients [[#References |[6]]]. Because E. nodatum is a periodontal pathogen, research has been focused on learning about oral microbiota, and how changes in these microbiota affect and are affected by different diseases. | ||
= | =Discovery= | ||
In the late 1980s, bacterial species that were involved in the initiation of periodontitis were discovered [[#References |[4]]]. The different bacterial species were separated into two categories, the red and the orange complex, depending on how severely they influence the progression of periodontitis. Some of the bacteria within the orange complex are: Streptococcus constellatus, Prevotella intermedia and Eubacterium nodatum [[#References |[7]]]. Periodontal disease was first discovered in mummies in ancient Egypt in the 1700s [[#References |[8]]]. | In the late 1980s, bacterial species that were involved in the initiation of periodontitis were discovered [[#References |[4]]]. The different bacterial species were separated into two categories, the red and the orange complex, depending on how severely they influence the progression of periodontitis. Some of the bacteria within the orange complex are: Streptococcus constellatus, Prevotella intermedia and Eubacterium nodatum [[#References |[7]]]. Periodontal disease was first discovered in mummies in ancient Egypt in the 1700s [[#References |[8]]]. | ||
= | = Genome structure= | ||
There are a total of 1.83 million base pairs that make up the E. nodatum genome [[#References |[1]]]. E. nodatum has a total of 1661 genes and 1543 of which code for proteins involved in amino acid adenylation, antiporter transportation and proteins which play the role of kinases, transferases, polymerases, and transcription and translation factors [[#References |[1]]]. The genome has a guanine-cytosine percentage of 38.1 [[#References |[1]]]. The genetic sequence of E. nodatum is 93.6% similar to Eubacterium tardum, a species in the same genius [[#References |[9]]]. | There are a total of 1.83 million base pairs that make up the E. nodatum genome [[#References |[1]]]. E. nodatum has a total of 1661 genes and 1543 of which code for proteins involved in amino acid adenylation, antiporter transportation and proteins which play the role of kinases, transferases, polymerases, and transcription and translation factors [[#References |[1]]]. The genome has a guanine-cytosine percentage of 38.1 [[#References |[1]]]. The genetic sequence of E. nodatum is 93.6% similar to Eubacterium tardum, a species in the same genius [[#References |[9]]]. | ||
= | =Cell Structure and Metabolic Process= | ||
E. nodatum are branched, filamentous, Gram-positive bacteria that are nonmotile, and do not produce spores [[#References |[3]]]. When colonized, the rod-shaped bacteria clump together in broth cultures [Figure 2]. When they form these clumps under favorable conditions, a biofilm is created [[#References |[3]]]. After incubation, cells form circular and raspberry-shaped colonies that are cream-colored [[#References |[3]]]. [[#References |[10]]]. | E. nodatum are branched, filamentous, Gram-positive bacteria that are nonmotile, and do not produce spores [[#References |[3]]]. When colonized, the rod-shaped bacteria clump together in broth cultures [Figure 2]. When they form these clumps under favorable conditions, a biofilm is created [[#References |[3]]]. After incubation, cells form circular and raspberry-shaped colonies that are cream-colored [[#References |[3]]]. [[#References |[10]]]. | ||
[[File:Figure_2.jpeg|200px|thumb|right|Figure 2. Microscopic image of E. Nodatum; rod-shaped and Gram-positive bacteria that clumps together. | |||
Source: http://phil.cdc.gov/phil/details.asp?pid=11195]] | |||
E. nodatum are obligately anaerobic, non-saccharolytic, grow optimally at 37 ºC, and can produce butyrate and acetate [[#References |[4]]]. E. nodatum can also hydrolyze arginine and lysine, and thus use these as substrates for energy sources [[#References |[4]]]. | E. nodatum are obligately anaerobic, non-saccharolytic, grow optimally at 37 ºC, and can produce butyrate and acetate [[#References |[4]]]. E. nodatum can also hydrolyze arginine and lysine, and thus use these as substrates for energy sources [[#References |[4]]]. | ||
= | =Ecology= | ||
E. nodatum is characterized as an opportunistic pathogen that can adhere to inanimate objects[[#References |[11]]]. E. nodatum is commonly found in anaerobic environments such as the peri-implant sulcus, and the female reproductive tract [[#References |[ | E. nodatum is characterized as an opportunistic pathogen that can adhere to inanimate objects [[#References |[11]]]. E. nodatum is commonly found in anaerobic environments such as the peri-implant sulcus, and the female reproductive tract [[#References |[11]]] [[#References |[12]]]. Additionally, E. nodatum has been found benignly in the head and the neck [[#References |[10]]]. | ||
= | =Pathology= | ||
The primary pathogenesis of E. nodatum is periodontitis. Periodontitis is the result of a biofilm created by microorganisms that are part of the orange complex [[#References |[13]]]. Surgical orthodontic treatments that utilize fixed orthodontic appliances cause changes in periodontal bacteria, like E. nodatum, leading to a buildup of plaque, as shown in Figure 3, and can increase a patient’s risk of periodontal disease [[#References |[14]]]. When fixed orthodontic appliances are removed, plaque levels decline [[#References |[14]]]. | The primary pathogenesis of E. nodatum is periodontitis. Periodontitis is the result of a biofilm created by microorganisms that are part of the orange complex [[#References |[13]]]. Surgical orthodontic treatments that utilize fixed orthodontic appliances cause changes in periodontal bacteria, like E. nodatum, leading to a buildup of plaque, as shown in Figure 3, and can increase a patient’s risk of periodontal disease [[#References |[14]]]. When fixed orthodontic appliances are removed, plaque levels decline [[#References |[14]]]. | ||
[[File: Figure_3.jpeg|200px|thumb|left|Figure 3. Image of bacteria developing in the human oral cavity on dental surfaces. | |||
Source: https://biotop.boku.ac.at/projects/Schaeffer3.html]] | |||
E. nodatum causes pelvic inflammatory disease when it colonizes the female genital tract. In the 1980s, before aseptic techniques were practiced, E. nodatum colonized the female genital tract through contaminated intrauterine devices for contraception, eventually causing pelvic inflammatory disease [[#References |[11]]]. E. nodatum is very similar to Actinomyces israelii in its morphology as well as its colonizing strategies [[#References |[11]]]. A. israelii commonly infects livestock with a disease called lumpy jaw, but rarely infects humans [[#References |[15]]]. E. nodatum infects humans through the same strategy causing severe periodontitis [[#References |[11]]]. It is generally unknown the pathologic biomechanisms used by E. nodatum in relation to other microorganisms in the biofilm due to their uncultivable nature [[#References |[14]]]. | |||
=Current Research= | |||
= | Current research is concentrated on exploring the pathogenesis of E. nodatum in the context of periodontitis [[#References |[14]]] [[#References |[15]]] [[#References |[16]]] Smoking is an amplifier for E. nodatum; research has found that after scaling and root planing in individuals with chronic periodontitis, E. nodatum re-colonizes more in smokers than their nonsmoking counterparts [[#References |[2]]]. It is generally unknown the pathologic biomechanisms used by E. nodatum in relation to other microorganisms in the biofilm due to their uncultivable nature [[#References |[14]]]. | ||
Current research is concentrated on exploring the pathogenesis of E. nodatum in the context of periodontitis [[#References |[14]]] | |||
There is also ongoing research centered around discovering oral pathogens similar to E. nodatum that are active in the peri-implantitis of oral implants [[#References |[7]]]. Similarly, studies are being conducted to explore new oral pathogens involved in chronic periodontitis [[#References |[15]]]. Other research projects are currently investigating associations between periodontal bacteria and stroke, cognitive impairment, and Alzheimer’s disease [[#References |[5]]]. Risk for developing Alzheimer's disease has been correlated with serum levels of common periodontal microbiota, such as E. nodatum [[#References |[5]]]. | There is also ongoing research centered around discovering oral pathogens similar to E. nodatum that are active in the peri-implantitis of oral implants [[#References |[7]]]. Similarly, studies are being conducted to explore new oral pathogens involved in chronic periodontitis [[#References |[15]]]. Other research projects are currently investigating associations between periodontal bacteria and stroke, cognitive impairment, and Alzheimer’s disease [[#References |[5]]]. Risk for developing Alzheimer's disease has been correlated with serum levels of common periodontal microbiota, such as E. nodatum [[#References |[5]]]. | ||
| Line 30: | Line 39: | ||
The effects of environmental and systemic factors in diabetes mellitus and smoking on chronic periodontitis pathogenic bacterial species including E. nodatum are also actively being studied in order to develop effective therapeutic treatments for the disease [[#References |[10]]]. | The effects of environmental and systemic factors in diabetes mellitus and smoking on chronic periodontitis pathogenic bacterial species including E. nodatum are also actively being studied in order to develop effective therapeutic treatments for the disease [[#References |[10]]]. | ||
= | |||
=References= | |||
<br>[https://www.ncbi.nlm.nih.gov/genome/13125[1]] Eubacterium nodatum (ID 13125) - Genome - NCBI. (n.d.). Retrieved October 24, 2016. Web. | <br>[https://www.ncbi.nlm.nih.gov/genome/13125[1]] Eubacterium nodatum (ID 13125) - Genome - NCBI. (n.d.). Retrieved October 24, 2016. Web. | ||
<br>[https://doi.org/10.11607/jomi.2570[2]] Tamura, N., Ochi, M., Miyakawa, H., & Nakazawa, F. 2013. Analysis of bacterial flora associated with peri-implantitis using obligate anaerobic culture technique and 16S rDNA gene sequence. The International Journal of Oral & Maxillofacial Implants,28(6), 1521–1529. Web. | |||
associated with peri-implantitis using obligate anaerobic culture technique and 16S | |||
rDNA gene sequence. The International Journal of Oral & Maxillofacial Implants, | |||
28(6), 1521–1529.. | |||
[ | <br>[http://www.microbiologyresearch.org/docserver/fulltext/ijsem/46/1/ijs-46-1-31.pdf?expires=1481567902&id=id&accname=guest&checksum=E4A5825C8DE1B5100F4F952993A3F108[3]] Holdeman, L. V., Cato, E. P., Burmeister, J. A., & Moore, C.E. 1980. Descriptions of Eubacterium timidum sp. nov., Eubacterium branchy sp. nov., and Eubacterium nodatum sp. nov. Isolated from human Periodontitis. International Journal of Systematic Bacteriology, 30(1), 163-169. Web. | ||
[ | <br>[http://www.tandfonline.com/doi/abs/10.3109/08910609609166471[4]] Uematsu, H., & Hoshino, E. 1996. Degradation of arginine and other amino acids by Eubacterium nodatum ATCC 33099. Microbial Ecology in Health and Disease, 9, 305-311. Web. | ||
<br>[https://doi.org/10.1371/journal.pone.0114959[5]] Noble, J. M., Scarmeas, N., Celenti, R. S., Elkind, M. S. V., Wright, C. B., Schupf, N., & Papapanou, P. N. 2014. Serum IgG antibody levels to periodontal microbiota are associated with incident Alzheimer disease. PLoS ONE, 9(12). Web. | |||
<br>[https://doi.org/10.1177/0022034512457373[6]] Eke, P. I., Dye, B. A., Wei, L., Thornton-Evans, G. O., & Genco, R. J. 2012. Prevalence of periodontitis in adults in the United States: 2009 and 2010. Journal of Dental Research, 91(10), 914–920. Web. | |||
[ | <br>[https://doi.org/10.1111/j.1600-051X.1998.tb02419.x[7]] Socransky, S. S., Haffajee, A. D., Cugini, M. A., Smith, C., & Kent, R. L. 1998. Microbial complexes in subgingival plaque. Journal of Clinical Periodontology, 25(2), 134–144. Web. | ||
[ | <br>[https://doi.org/10.1038/sj.bdj.2009.309[8]] Forshaw, R. J. 2009. Dental health and disease in ancient Egypt. British Dental Journal, 206(8), 421–424. Web. | ||
[ | <br>[http://cid.oxfordjournals.org/content/25/Supplement_2/S78.full.pdf[9]] Jousimies-Somer, H. 1997. Recently described clinically important anaerobic bacteria: taxonomic aspects and update. Clinical Infectious Diseases, 25(Suppl 2), S78–87. Web. | ||
[ | <br>[http://jcm.asm.org/content/25/8/1540[10]] Hill, G. B., Ayers, O. M., & Kohan, A. P. 1987. Characteristics and sites of infection of Eubacterium nodatum, Eubacterium timidum, Eubacterium brachy, and other asaccharolytic eubacteria. Journal of Clinical Microbiology, 25(8), 1540–1545. Web. | ||
<br>[https://www.ncbi.nlm.nih.gov/pubmed/1553171[11]] Hill, G. B. 1992. Eubacterium nodatum mimics Actinomyces in intrauterine device-associated infections and other settings within the female genital tract. Obstetrics & Gynecology, 79(4), 534-538. Web. | |||
[ | <br>[https://doi.org/10.11607/jomi.2570[12]] Tamura, N., Ochi, M., Miyakawa, H., & Nakazawa, F. 2013. Analysis of Bacterial Flora Associated with Peri-implantitis Using Obligate Anaerobic Culture Technique and 16S rDNA Gene Sequence. The International Journal of Oral & Maxillofacial Implants, 28(6), 1521–1529. Web. | ||
<br>[https://microbewiki.kenyon.edu/index.php/Prevotella_nigrescens[13]] Prevotella nigrescens - MicrobeWiki. (n.d.). Retrieved September 20, 2016. Web. | |||
[ | <br>[http://onlinelibrary.wiley.com/doi/10.1111/ocr.12037/abstract[14]] Ireland, A. J., V. Soro, S. V. Sprague, N. W. T. Harradine, C. Day, S. Al-Anezi, H. F. Jenkinson, M. Sherriff, D. Dymock, and J. R. Sandy. 2013. The effects of different orthodontic appliances upon microbial communities. Orthodontics & Craniofacial Research 17, 115-123. Web. | ||
<br>[https://doi.org/10.2147/IDR.S39601[15]] Valour, F., Sénéchal, A., Dupieux, C., Karsenty, J., Lustig, S., Breton, P., … Ferry, T. 2014. Actinomycosis: etiology, clinical features, diagnosis, treatment, and management. Infection and Drug Resistance, 7, 183–197. Web. | |||
<br>[https://doi.org/10.1034/j.1399-302X.1999.140107.x[16]] Spratt, D. A., Weightman, A. J., & Wade, W. G. 1999. Diversity of oral asaccharolytic Eubacterium species in periodontitis – identification of novel phylotypes representing uncultivated taxa. Oral Microbiology and Immunology, 14(1), 56–59. Web. | |||
<br>[https://doi.org/10.1111/adj.12225[17]] Feres, M., Bernal, M., Matarazzo, F., Faveri, M., Duarte, P., & Figueiredo, L. 2015. Subgingival bacterial recolonization after scaling and root planing in smokers with chronic periodontitis. Australian Dental Journal, 60(2), 225–232. .(1998). Web. | |||
<br><br> | <br><br> | ||
<br>Edited by [ | <br>Edited by [Ronald Akiki, Jessica Bonet, Monica Eng, and Taylor Juretic], students of [mailto:jmtalbot@bu.edu Jennifer Talbot] for [http://www.bu.edu/academics/cas/courses/cas-bi-311/ BI 311 General Microbiology], 2016, [http://www.bu.edu/ Boston University]. | ||
[[Category:Pages edited by students of Jennifer Talbot at Boston University]] | [[Category:Pages edited by students of Jennifer Talbot at Boston University]] | ||
Latest revision as of 18:26, 12 December 2016
Classification
a. Higher order taxa
Bacteria, Firmicutes, Clostridia, Clostridiale, Clostridiales Family XIII, Eubacterium [1]
Description and significance
Eubacterium nodatum is one of the predominant bacteria found in peri-implant sites [2]. E. nodatum is a species of Eubacterium [Figure 1] that has commonly been found in patients with chronic periodontal disease. E. nodatum is obligately anaerobic, and a Gram-positive microbe [3]. Although this bacterium was initially identified in the late 1980s, it has been difficult to understand its metabolic pathways since it is challenging to isolate, has a slow growth process, and is structurally complex [4]. [5]. According to the Centers for Disease Control and Prevention (CDC), 47.2% of American adults suffer from periodontitis, a bacterial infection that forms on oral surfaces and damages the tissues that surround teeth in peri-implant patients [6]. Because E. nodatum is a periodontal pathogen, research has been focused on learning about oral microbiota, and how changes in these microbiota affect and are affected by different diseases.
Discovery
In the late 1980s, bacterial species that were involved in the initiation of periodontitis were discovered [4]. The different bacterial species were separated into two categories, the red and the orange complex, depending on how severely they influence the progression of periodontitis. Some of the bacteria within the orange complex are: Streptococcus constellatus, Prevotella intermedia and Eubacterium nodatum [7]. Periodontal disease was first discovered in mummies in ancient Egypt in the 1700s [8].
Genome structure
There are a total of 1.83 million base pairs that make up the E. nodatum genome [1]. E. nodatum has a total of 1661 genes and 1543 of which code for proteins involved in amino acid adenylation, antiporter transportation and proteins which play the role of kinases, transferases, polymerases, and transcription and translation factors [1]. The genome has a guanine-cytosine percentage of 38.1 [1]. The genetic sequence of E. nodatum is 93.6% similar to Eubacterium tardum, a species in the same genius [9].
Cell Structure and Metabolic Process
E. nodatum are branched, filamentous, Gram-positive bacteria that are nonmotile, and do not produce spores [3]. When colonized, the rod-shaped bacteria clump together in broth cultures [Figure 2]. When they form these clumps under favorable conditions, a biofilm is created [3]. After incubation, cells form circular and raspberry-shaped colonies that are cream-colored [3]. [10].
E. nodatum are obligately anaerobic, non-saccharolytic, grow optimally at 37 ºC, and can produce butyrate and acetate [4]. E. nodatum can also hydrolyze arginine and lysine, and thus use these as substrates for energy sources [4].
Ecology
E. nodatum is characterized as an opportunistic pathogen that can adhere to inanimate objects [11]. E. nodatum is commonly found in anaerobic environments such as the peri-implant sulcus, and the female reproductive tract [11] [12]. Additionally, E. nodatum has been found benignly in the head and the neck [10].
Pathology
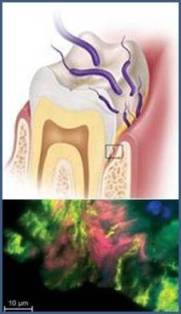
The primary pathogenesis of E. nodatum is periodontitis. Periodontitis is the result of a biofilm created by microorganisms that are part of the orange complex [13]. Surgical orthodontic treatments that utilize fixed orthodontic appliances cause changes in periodontal bacteria, like E. nodatum, leading to a buildup of plaque, as shown in Figure 3, and can increase a patient’s risk of periodontal disease [14]. When fixed orthodontic appliances are removed, plaque levels decline [14].
E. nodatum causes pelvic inflammatory disease when it colonizes the female genital tract. In the 1980s, before aseptic techniques were practiced, E. nodatum colonized the female genital tract through contaminated intrauterine devices for contraception, eventually causing pelvic inflammatory disease [11]. E. nodatum is very similar to Actinomyces israelii in its morphology as well as its colonizing strategies [11]. A. israelii commonly infects livestock with a disease called lumpy jaw, but rarely infects humans [15]. E. nodatum infects humans through the same strategy causing severe periodontitis [11]. It is generally unknown the pathologic biomechanisms used by E. nodatum in relation to other microorganisms in the biofilm due to their uncultivable nature [14].
Current Research
Current research is concentrated on exploring the pathogenesis of E. nodatum in the context of periodontitis [14] [15] [16] Smoking is an amplifier for E. nodatum; research has found that after scaling and root planing in individuals with chronic periodontitis, E. nodatum re-colonizes more in smokers than their nonsmoking counterparts [2]. It is generally unknown the pathologic biomechanisms used by E. nodatum in relation to other microorganisms in the biofilm due to their uncultivable nature [14]. There is also ongoing research centered around discovering oral pathogens similar to E. nodatum that are active in the peri-implantitis of oral implants [7]. Similarly, studies are being conducted to explore new oral pathogens involved in chronic periodontitis [15]. Other research projects are currently investigating associations between periodontal bacteria and stroke, cognitive impairment, and Alzheimer’s disease [5]. Risk for developing Alzheimer's disease has been correlated with serum levels of common periodontal microbiota, such as E. nodatum [5].
Inter-bacterial relationships are being studied more closely in oral ecology with the goal of shifting the focus from the removal of pathogenic oral bacteria to reestablishing the optimal colonization balance in oral plaques [16]. The species that are vigorously being studied (other than E. nodatum) include Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola, Porphyromonas micra and Porphyromonas intermedia [16]. There is also a focus on research regarding orthodontic treatment in order to develop better equipment to lower the prevalence of negative outcomes, such as plaque buildup, from orthodontic interventions [10].
The effects of environmental and systemic factors in diabetes mellitus and smoking on chronic periodontitis pathogenic bacterial species including E. nodatum are also actively being studied in order to develop effective therapeutic treatments for the disease [10].
References
[1] Eubacterium nodatum (ID 13125) - Genome - NCBI. (n.d.). Retrieved October 24, 2016. Web.
[2] Tamura, N., Ochi, M., Miyakawa, H., & Nakazawa, F. 2013. Analysis of bacterial flora associated with peri-implantitis using obligate anaerobic culture technique and 16S rDNA gene sequence. The International Journal of Oral & Maxillofacial Implants,28(6), 1521–1529. Web.
[3] Holdeman, L. V., Cato, E. P., Burmeister, J. A., & Moore, C.E. 1980. Descriptions of Eubacterium timidum sp. nov., Eubacterium branchy sp. nov., and Eubacterium nodatum sp. nov. Isolated from human Periodontitis. International Journal of Systematic Bacteriology, 30(1), 163-169. Web.
[4] Uematsu, H., & Hoshino, E. 1996. Degradation of arginine and other amino acids by Eubacterium nodatum ATCC 33099. Microbial Ecology in Health and Disease, 9, 305-311. Web.
[5] Noble, J. M., Scarmeas, N., Celenti, R. S., Elkind, M. S. V., Wright, C. B., Schupf, N., & Papapanou, P. N. 2014. Serum IgG antibody levels to periodontal microbiota are associated with incident Alzheimer disease. PLoS ONE, 9(12). Web.
[6] Eke, P. I., Dye, B. A., Wei, L., Thornton-Evans, G. O., & Genco, R. J. 2012. Prevalence of periodontitis in adults in the United States: 2009 and 2010. Journal of Dental Research, 91(10), 914–920. Web.
[7] Socransky, S. S., Haffajee, A. D., Cugini, M. A., Smith, C., & Kent, R. L. 1998. Microbial complexes in subgingival plaque. Journal of Clinical Periodontology, 25(2), 134–144. Web.
[8] Forshaw, R. J. 2009. Dental health and disease in ancient Egypt. British Dental Journal, 206(8), 421–424. Web.
[9] Jousimies-Somer, H. 1997. Recently described clinically important anaerobic bacteria: taxonomic aspects and update. Clinical Infectious Diseases, 25(Suppl 2), S78–87. Web.
[10] Hill, G. B., Ayers, O. M., & Kohan, A. P. 1987. Characteristics and sites of infection of Eubacterium nodatum, Eubacterium timidum, Eubacterium brachy, and other asaccharolytic eubacteria. Journal of Clinical Microbiology, 25(8), 1540–1545. Web.
[11] Hill, G. B. 1992. Eubacterium nodatum mimics Actinomyces in intrauterine device-associated infections and other settings within the female genital tract. Obstetrics & Gynecology, 79(4), 534-538. Web.
[12] Tamura, N., Ochi, M., Miyakawa, H., & Nakazawa, F. 2013. Analysis of Bacterial Flora Associated with Peri-implantitis Using Obligate Anaerobic Culture Technique and 16S rDNA Gene Sequence. The International Journal of Oral & Maxillofacial Implants, 28(6), 1521–1529. Web.
[13] Prevotella nigrescens - MicrobeWiki. (n.d.). Retrieved September 20, 2016. Web.
[14] Ireland, A. J., V. Soro, S. V. Sprague, N. W. T. Harradine, C. Day, S. Al-Anezi, H. F. Jenkinson, M. Sherriff, D. Dymock, and J. R. Sandy. 2013. The effects of different orthodontic appliances upon microbial communities. Orthodontics & Craniofacial Research 17, 115-123. Web.
[15] Valour, F., Sénéchal, A., Dupieux, C., Karsenty, J., Lustig, S., Breton, P., … Ferry, T. 2014. Actinomycosis: etiology, clinical features, diagnosis, treatment, and management. Infection and Drug Resistance, 7, 183–197. Web.
[16] Spratt, D. A., Weightman, A. J., & Wade, W. G. 1999. Diversity of oral asaccharolytic Eubacterium species in periodontitis – identification of novel phylotypes representing uncultivated taxa. Oral Microbiology and Immunology, 14(1), 56–59. Web.
[17] Feres, M., Bernal, M., Matarazzo, F., Faveri, M., Duarte, P., & Figueiredo, L. 2015. Subgingival bacterial recolonization after scaling and root planing in smokers with chronic periodontitis. Australian Dental Journal, 60(2), 225–232. .(1998). Web.
Edited by [Ronald Akiki, Jessica Bonet, Monica Eng, and Taylor Juretic], students of Jennifer Talbot for BI 311 General Microbiology, 2016, Boston University.
