Vibrio alginolyticus: Difference between revisions
No edit summary |
No edit summary |
||
| (One intermediate revision by the same user not shown) | |||
| Line 19: | Line 19: | ||
Organisms from the ''Vibrio genus'' were first observed by Filippo Pacini in 1854, and were further described by Robert Koch in 1883. The species ''V. alginolyticus'' emerged on the scene as a result of the need to further classify another ''Vibrio species'', ''Vibrio parahaemolyticus''. ''V. parahaemolyticus'' historically had included two biotypes, but it was suggested by Zen-Yoji et al. in 1965 the second biotype be moved into its own species. In 1968, Sakazaki et al. proposed a new name for the biotype species: ''Vibrio alginolyticus''. ''V. alginolyticus'' would later be isolated in 1997, as a result of a pathogenic outbreak in Russia.<sup>9, | Organisms from the ''Vibrio genus'' were first observed by Filippo Pacini in 1854, and were further described by Robert Koch in 1883. The species ''V. alginolyticus'' emerged on the scene as a result of the need to further classify another ''Vibrio species'', ''Vibrio parahaemolyticus''. ''V. parahaemolyticus'' historically had included two biotypes, but it was suggested by Zen-Yoji et al. in 1965 the second biotype be moved into its own species. In 1968, Sakazaki et al. proposed a new name for the biotype species: ''Vibrio alginolyticus''. ''V. alginolyticus'' would later be isolated in 1997, as a result of a pathogenic outbreak in Russia.<sup>9, 12</sup> | ||
| Line 28: | Line 28: | ||
==Genome and | ==Genome and Genetics== | ||
''Vibrio'' organisms belong to the larger prokaryotic branch of ''Proteobacteria''. ''V. alginolyticus'' itself is closely related to the ''Vibrio'' species of ''V. cambellii'', ''V. parahaemolyticus'', and ''V. proteolyticus'', based on 16S rRNA sequencing.<sup>2</sup> | ''Vibrio'' organisms belong to the larger prokaryotic branch of ''Proteobacteria''. ''V. alginolyticus'' itself is closely related to the ''Vibrio'' species of ''V. cambellii'', ''V. parahaemolyticus'', and ''V. proteolyticus'', based on 16S rRNA sequencing.<sup>2</sup> | ||
| Line 35: | Line 35: | ||
==Nutrition and | ==Nutrition and Metabolism== | ||
While ''Vibrio'' species do vary greatly in their nutrition and growth requirements, one feature shared by all members of this genus is that they are halophilic, requiring the presence of Na<sup>+</sup> to grow.<sup>2</sup> ''V. alginolyticus'' has an absolute salt requirement, meaning it will not grow unless in the presence of salt. For ''V. alginolyticus'' the minimal concentration of NaCl required for optimal growth is approximately 200-260mM.<sup>2</sup> Most ''Vibrio'' species prefer a pH range of 7-8 for optimal growth, at a temperature of 18-22 °C. | While ''Vibrio'' species do vary greatly in their nutrition and growth requirements, one feature shared by all members of this genus is that they are halophilic, requiring the presence of Na<sup>+</sup> to grow.<sup>2</sup> ''V. alginolyticus'' has an absolute salt requirement, meaning it will not grow unless in the presence of salt. For ''V. alginolyticus'' the minimal concentration of NaCl required for optimal growth is approximately 200-260mM.<sup>2</sup> Most ''Vibrio'' species prefer a pH range of 7-8 for optimal growth, at a temperature of 18-22 °C. | ||
| Line 56: | Line 56: | ||
==Current Research== | ==Current Research== | ||
The complete genome sequence of ''Vibrio alginolyticus''' was published late 2016,<sup>4</sup> due primarily to the need for further research into the bacteria. ''V. alginolyticus'' is responsible for significant mortality and economic losses in many commercial fish, and the researchers responsible for the genome sequencing wanted to respond to the threat posed by ''V. alginolyticus''. By sequencing the complete genome, the researchers were able to identify many coding, non-coding and pseudogenes, which the researchers hope will contribute to further study of the bacteria, as well as prove useful for comparing ''V. aglinolyticus'' to other vibrios. | |||
In addition to genome sequencing, another article published in 2012<sup>11</sup> revealed the prevelance of ''V. alginolyticus'' in Bangladesh. The researchers took sediment samples from three costal areas and six rivers in Bangladesh, and, after characterization and differential tests, were found to contain ''V. alginolyticus'' cells. These cells were tested for antibiotic susceptibility, and their effect on a species of fish. It was found that ''V. alginolyticus'' is only susceptible to one of the six commonly used antibiotics tested, nalidixic acid, and resistant to the others. It was noted that the mortality of the fish increased with increased concentration of ''V. alginolyticus'', where infected fish were able to infect healthy fish. The researchers argued that one of the virulence factors for ''V. alginolyticus'' might be an extracellular toxin, and proposed that a characterization of this toxin could be a potential start for further pathogenicity research. | |||
==References== | ==References== | ||
1. Balebona MC, Andreu MJ, Bordas MA, Zorrilla I, Moriñigo MA, Borrego JJ. Pathogenicity of Vibrio alginolyticus for Cultured Gilt-Head Sea Bream (Sparus aurata L.). Applied and Environmental Microbiology. 1998;64(11):4269-4275. | 1. Balebona MC, Andreu MJ, Bordas MA, Zorrilla I, Moriñigo MA, Borrego JJ. Pathogenicity of ''Vibrio alginolyticus'' for Cultured Gilt-Head Sea Bream (Sparus aurata L.). Applied and Environmental Microbiology. 1998;64(11):4269-4275. | ||
| Line 69: | Line 73: | ||
4. Deng Y, Chen C, Zhao Z, Huang X, Yang Y, Ding X. Complete Genome Sequence of Vibrio alginolyticus ZJ-T. Genome Announcements. 2016;4(5):e00912-16. doi:10.1128/genomeA.00912-16. | 4. Deng Y, Chen C, Zhao Z, Huang X, Yang Y, Ding X. Complete Genome Sequence of ''Vibrio alginolyticus'' ZJ-T. Genome Announcements. 2016;4(5):e00912-16. doi:10.1128/genomeA.00912-16. | ||
5. Deng Y, Chen C, Zhao Z, Zhao J, Jacq A, Huang X, Yany Y. The RNA Chaperone Hfq Is Involved in Colony Morphology, Nutrient Utilization and Oxidative and Envelope Stress Response in Vibrio alginolyticus. Skurnik M, ed. PLoS ONE. 2016;11(9):e0163689. | 5. Deng Y, Chen C, Zhao Z, Zhao J, Jacq A, Huang X, Yany Y. The RNA Chaperone Hfq Is Involved in Colony Morphology, Nutrient Utilization and Oxidative and Envelope Stress Response in ''Vibrio alginolyticus''. Skurnik M, ed. PLoS ONE. 2016;11(9):e0163689. | ||
6. Gomez JM, Fajardo R, Patiño JF, Arias CA. Necrotizing Fasciitis Due to Vibrio alginolyticus in an Immunocompetent Patient. Journal of Clinical Microbiology. 2003;41(7):3427-3429. | 6. Gomez JM, Fajardo R, Patiño JF, Arias CA. Necrotizing Fasciitis Due to ''Vibrio alginolyticus'' in an Immunocompetent Patient. Journal of Clinical Microbiology. 2003;41(7):3427-3429. | ||
| Line 81: | Line 85: | ||
8. Martins ML, Mouriño JLP, Fezer GF, Buglione Neto CC, Garcia P, Silva BC, Jatobá A, Vieira FN. Isolation and experimental infection with Vibrio alginolyticus in the sea horse, Hippocampus reidi Ginsburg, 1933 (Osteichthyes: Syngnathidae) in Brazil. Brazilian Journal of Biology. 2010;70(1): 205-209. | 8. Martins ML, Mouriño JLP, Fezer GF, Buglione Neto CC, Garcia P, Silva BC, Jatobá A, Vieira FN. Isolation and experimental infection with ''Vibrio alginolyticus'' in the sea horse, ''Hippocampus reidi'' Ginsburg, 1933 (Osteichthyes: Syngnathidae) in Brazil. Brazilian Journal of Biology. 2010;70(1): 205-209. | ||
| Line 87: | Line 91: | ||
10. Sakazaki R. Proposal of Vibrio alginolyticus for the biotype 2 of Vibrio parahaemolyticus. Japanese Journal of Medical Science and Biology. 1968;21(5):359–62. | 10. Sakazaki R. Proposal of ''Vibrio alginolyticus'' for the biotype 2 of Vibrio parahaemolyticus. Japanese Journal of Medical Science and Biology. 1968;21(5):359–62. | ||
11. Siddiqui1 R, Alam MM, Naser MN, Otomo Y, Yasmin M, Nessa J, Ahsan CR. Prevalence of ''Vibrio alginolyticus'' in Sediment Samples of River and Coastal Areas of Bangladesh. Bangladesh J Microbiol. 2012;29(1):1-6. | |||
12. Smolikova LM, Lomov LM, Khomenko TV, Murnachev GP, Kudriakova TA, Fetsailova OP, Sanamiants EM, Makedonova LD, Kachkina GV, | |||
Golenishcheva EN. Studies on halophilic vibrios causing a food poisoning outbreak in the city of Vladivostok. Zh Mikrobiol Epidemiol Immunobiol. 2001;6:3-7. | Golenishcheva EN. Studies on halophilic vibrios causing a food poisoning outbreak in the city of Vladivostok. Zh Mikrobiol Epidemiol Immunobiol. 2001;6:3-7. | ||
Latest revision as of 21:13, 31 March 2017
Classification
Higher order taxa
Bacteria; Proteobacteria; Gammaproteobacteria; Vibrionales; Vibrionaceae; Vibrio
Species
|
NCBI: Taxonomy |
Vibrio alginolyticus
Description and significance
Organisms from the Vibrio genus were first observed by Filippo Pacini in 1854, and were further described by Robert Koch in 1883. The species V. alginolyticus emerged on the scene as a result of the need to further classify another Vibrio species, Vibrio parahaemolyticus. V. parahaemolyticus historically had included two biotypes, but it was suggested by Zen-Yoji et al. in 1965 the second biotype be moved into its own species. In 1968, Sakazaki et al. proposed a new name for the biotype species: Vibrio alginolyticus. V. alginolyticus would later be isolated in 1997, as a result of a pathogenic outbreak in Russia.9, 12
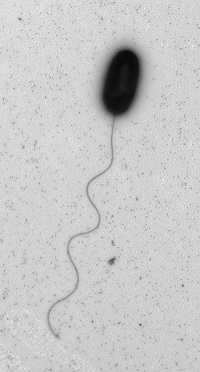
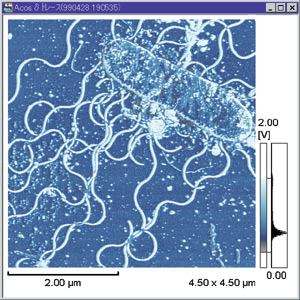
V. alginolyticus can be “comma” or coccoid shaped and is Gram negative.7 There are a few different strains, many of which are pathogenic. V. alginolyticus displays a single polar flagella in liquid media, but when the bacteria is introduced to solid media, morphogenetic changes occur to produce numerous lateral flagella (see photo).2
(For more information, see: Vibrio).
Genome and Genetics
Vibrio organisms belong to the larger prokaryotic branch of Proteobacteria. V. alginolyticus itself is closely related to the Vibrio species of V. cambellii, V. parahaemolyticus, and V. proteolyticus, based on 16S rRNA sequencing.2
The complete genome of V. alginolyticus has been sequenced by Illumina high-throughput sequencing.4 The genome was revealed to have a total size of 5,406,094 bp and G+C content of 44.71%. The genome has 4,644 coding genes present. The complete sequence was deposited at GeneBank under the ascension numbers CP016224 and CP016225.
Nutrition and Metabolism
While Vibrio species do vary greatly in their nutrition and growth requirements, one feature shared by all members of this genus is that they are halophilic, requiring the presence of Na+ to grow.2 V. alginolyticus has an absolute salt requirement, meaning it will not grow unless in the presence of salt. For V. alginolyticus the minimal concentration of NaCl required for optimal growth is approximately 200-260mM.2 Most Vibrio species prefer a pH range of 7-8 for optimal growth, at a temperature of 18-22 °C.
Vibrio alginolyticus was originally a biotype of V. parahaemolyticus, due to many similarities, but the differentiating factor was V. alginolyticus' ability to ferment sucrose.10 The bacteria is able to utilize citrate and other sugars as a carbon source, and also can produce extracellular enzymes which degrade DNA, RNA, proteins, lipids, and a variety of carbohydrates.2 V. alginolyticus in particular has been shown to be able to utilize d-glucose, d-maltose, d-trehalose, d-fructose or N-acetyl-d-glucosamine as its sole carbon source.
Ecology / Pathology
Water (and its associated microorganisms, animals, and plants) is the natural habitat of most vibrios. Often vibrios are associated with shellfish and other marine fish. Vibrios are also commonly associated with various types of plankton. 2
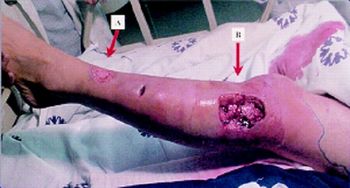
Vibrios are commonly studied for their pathogenicity. Numerous studies have been conducted, displaying the outbreak of V. alginolyticus in various marine animals. These studies show the necrotic effect of V. alginolyticus on different types of organisms; from fish to seahorses.1, 8 Though the exact pathogenic pathway is unknown,5 one study proposed a theoretical mechanism where V. alginolyticus adheres to mucosal layers of marine animals, where it is able to use the mucus for nutrients and causes ulcers and other tissue damage.1 V. alginolyticus is also known to cause necrosis of epithelial tissue in marine animals.8
Vibrio alginolyticus is also pathogenic to humans, known to cause gastroenteritis and other infections of the gastrointestinal tract.2 Additionally, V. alginolyticus has been isolated from soft tissue infections in humans, and there have been reported cases of humans contracting diseases as severe as necrotizing fasciitis following infection from V. alginolyticus, though most commonly the infection is gastroenteritis, otitis media, and otitis externa.3 Depending on the severity of the infection, simple antibiotics may be all that is required, and treatment tends to be focused more on the comfort of the patient, especially in the case of gastroenteritis. Cases are associated with the consumption of raw seafood or exposure of a wound to brackish water which may be housing V. alginolyticus.2
Current Research
The complete genome sequence of Vibrio alginolyticus' was published late 2016,4 due primarily to the need for further research into the bacteria. V. alginolyticus is responsible for significant mortality and economic losses in many commercial fish, and the researchers responsible for the genome sequencing wanted to respond to the threat posed by V. alginolyticus. By sequencing the complete genome, the researchers were able to identify many coding, non-coding and pseudogenes, which the researchers hope will contribute to further study of the bacteria, as well as prove useful for comparing V. aglinolyticus to other vibrios.
In addition to genome sequencing, another article published in 201211 revealed the prevelance of V. alginolyticus in Bangladesh. The researchers took sediment samples from three costal areas and six rivers in Bangladesh, and, after characterization and differential tests, were found to contain V. alginolyticus cells. These cells were tested for antibiotic susceptibility, and their effect on a species of fish. It was found that V. alginolyticus is only susceptible to one of the six commonly used antibiotics tested, nalidixic acid, and resistant to the others. It was noted that the mortality of the fish increased with increased concentration of V. alginolyticus, where infected fish were able to infect healthy fish. The researchers argued that one of the virulence factors for V. alginolyticus might be an extracellular toxin, and proposed that a characterization of this toxin could be a potential start for further pathogenicity research.
References
1. Balebona MC, Andreu MJ, Bordas MA, Zorrilla I, Moriñigo MA, Borrego JJ. Pathogenicity of Vibrio alginolyticus for Cultured Gilt-Head Sea Bream (Sparus aurata L.). Applied and Environmental Microbiology. 1998;64(11):4269-4275.
2. Bergey DH. Bergey's Manual of Systematic Bacteriology. Brenner D, Krieg N, Staley J, editors. 2nd ed. New York: Springer; 2005. Vol. 2, part B; pg. 494-546.
3. Carpenter C. Other pathogenic vibrios. Mandell GL, Bennett JE, Dolin R, editors. Mandell, Douglas and Bennett’s Principles and Practice of Infectious Disease. 4th ed. New York: Churchill Livingstone; 1995. Pg. 1945–8
4. Deng Y, Chen C, Zhao Z, Huang X, Yang Y, Ding X. Complete Genome Sequence of Vibrio alginolyticus ZJ-T. Genome Announcements. 2016;4(5):e00912-16. doi:10.1128/genomeA.00912-16.
5. Deng Y, Chen C, Zhao Z, Zhao J, Jacq A, Huang X, Yany Y. The RNA Chaperone Hfq Is Involved in Colony Morphology, Nutrient Utilization and Oxidative and Envelope Stress Response in Vibrio alginolyticus. Skurnik M, ed. PLoS ONE. 2016;11(9):e0163689.
6. Gomez JM, Fajardo R, Patiño JF, Arias CA. Necrotizing Fasciitis Due to Vibrio alginolyticus in an Immunocompetent Patient. Journal of Clinical Microbiology. 2003;41(7):3427-3429.
7. lbertini MC, Accorsi A, Teodori L, Pierfelici L, Uguccioni F, Rocchi MBL, Burattini S, Citterio B. Use of multiparameter analysis for Vibrio alginolyticus viable but nonculturable state determination. Cytometry. 2006;69A:260–265.
8. Martins ML, Mouriño JLP, Fezer GF, Buglione Neto CC, Garcia P, Silva BC, Jatobá A, Vieira FN. Isolation and experimental infection with Vibrio alginolyticus in the sea horse, Hippocampus reidi Ginsburg, 1933 (Osteichthyes: Syngnathidae) in Brazil. Brazilian Journal of Biology. 2010;70(1): 205-209.
9. Mustapha S, Mustapha EM, Nozha C. Vibrio Alginolyticus: an emerging pathogen of foodborne diseases. Int J Sci Technol. 2013;2(4):302-309.
10. Sakazaki R. Proposal of Vibrio alginolyticus for the biotype 2 of Vibrio parahaemolyticus. Japanese Journal of Medical Science and Biology. 1968;21(5):359–62.
11. Siddiqui1 R, Alam MM, Naser MN, Otomo Y, Yasmin M, Nessa J, Ahsan CR. Prevalence of Vibrio alginolyticus in Sediment Samples of River and Coastal Areas of Bangladesh. Bangladesh J Microbiol. 2012;29(1):1-6.
12. Smolikova LM, Lomov LM, Khomenko TV, Murnachev GP, Kudriakova TA, Fetsailova OP, Sanamiants EM, Makedonova LD, Kachkina GV,
Golenishcheva EN. Studies on halophilic vibrios causing a food poisoning outbreak in the city of Vladivostok. Zh Mikrobiol Epidemiol Immunobiol. 2001;6:3-7.
Authored by Parker Morris, a student of CJ Funk at John Brown University