Large Intestine: Difference between revisions
No edit summary |
|||
| (2 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | |||
==Introduction== | ==Introduction== | ||
According to the three-domain system, which is a biological classification scheme fabricated by Carl Woese, the cellular organisms that constitute life are divided into archaea, bacteria, and eukarya. Woese devised this categorization scheme by comparing the 16S rRNA sequences of living cells. The use of this specific ribosomal RNA was key to his success, as it was proved to be present in all living organisms. Therefore, comparison of this gene sequence was useful in determining the phylogeny of cellular life. This sequence differed between domains depending on the environment that surrounded the organisms as well as their method of metabolism. As a result, the prokaryotes were split into two domains, the archaea and bacteria, while the eukarya remained in a separate class due to their multicellular characteristics. [http://www.becomehealthynow.com/article/bodydigestive/787/ (1)] | According to the three-domain system, which is a biological classification scheme fabricated by Carl Woese, the cellular organisms that constitute life are divided into archaea, bacteria, and eukarya. Woese devised this categorization scheme by comparing the 16S rRNA sequences of living cells. The use of this specific ribosomal RNA was key to his success, as it was proved to be present in all living organisms. Therefore, comparison of this gene sequence was useful in determining the phylogeny of cellular life. This sequence differed between domains depending on the environment that surrounded the organisms as well as their method of metabolism. As a result, the prokaryotes were split into two domains, the archaea and bacteria, while the eukarya remained in a separate class due to their multicellular characteristics. [http://www.becomehealthynow.com/article/bodydigestive/787/ (1)] | ||
| Line 97: | Line 98: | ||
===Escherichia Coli=== | ===Escherichia Coli=== | ||
Common Species: ''Escherichia albertii; E. blattae; [[Escherichia | Common Species: ''Escherichia albertii; E. blattae; [[Escherichia coli|E. coli]]; E. fergusonii; E. hermannii; E. senegalensis; E. senegalensis; E. vulneris; E. sp.'' | ||
[[Escherichia coli]], also known as [[Escherichia coli|E. coli]], is an anaerobic gram-negative bacterium that can be found in large intestine of warm-blooded animals and humans. Even though it is a predominant and consistent organism, it contributes to only small portion of the content of total bacteria in GI tract. It is often used as an indicator to test the fecal contamination in nature such as water since [[Escherichia coli|E. coli]] is able to survive outside the body for a brief time period.(18)(19) Because of its simple nutritional requirement and ability to grow rapidly, [[Escherichia coli|E. coli]] is also useful in studying the organisms’ essential processes of life.(20) | [[Escherichia coli]], also known as [[Escherichia coli|E. coli]], is an anaerobic gram-negative bacterium that can be found in large intestine of warm-blooded animals and humans. Even though it is a predominant and consistent organism, it contributes to only small portion of the content of total bacteria in GI tract. It is often used as an indicator to test the fecal contamination in nature such as water since [[Escherichia coli|E. coli]] is able to survive outside the body for a brief time period.(18)(19) Because of its simple nutritional requirement and ability to grow rapidly, [[Escherichia coli|E. coli]] is also useful in studying the organisms’ essential processes of life.(20) | ||
| Line 270: | Line 271: | ||
Edited by | Edited by Benjamin Dae Lee, Hilary Otorowski, Julia Son, Rebecca Son, Alexander Pang, Ella Chen, David Araiza, students of [mailto:ralarsen@ucsd.edu Rachel Larsen] | ||
Latest revision as of 02:59, 20 August 2010
Introduction
According to the three-domain system, which is a biological classification scheme fabricated by Carl Woese, the cellular organisms that constitute life are divided into archaea, bacteria, and eukarya. Woese devised this categorization scheme by comparing the 16S rRNA sequences of living cells. The use of this specific ribosomal RNA was key to his success, as it was proved to be present in all living organisms. Therefore, comparison of this gene sequence was useful in determining the phylogeny of cellular life. This sequence differed between domains depending on the environment that surrounded the organisms as well as their method of metabolism. As a result, the prokaryotes were split into two domains, the archaea and bacteria, while the eukarya remained in a separate class due to their multicellular characteristics. (1)
Despite the common misconception that bacteria are organisms that only cause disease, they play an important role in facilitating our digestion. For example, the large intestine is home to hundreds of bacteria that aid in absorption, excretion, and catalysis of undigested foods. There are also bacteria present in the small intestine that support break down of foods passed down from the stomach as well as nutrient absorption. (2)
We will be focusing on prokaryotic, as well as eukaryotic, organisms that reside in the large intestine. The bacteria that will be discussed include the following: Lactobacillus, Bifidobacterium, Methanogens, Sulfate reducing bacteria, Bacteroides, Enterococcus, Escherichia coli, and Clostridium. In addition, we will discuss a couple of eukaryotic organisms, Candida and Entamoeba Histolytica, and see how it effects the large intestine.
Description of Niche
The large intestine, commonly known to be the final stage of digestion, is located in the abdominal cavity; specifically, between the small intestine and the anus. The primary functions of the large intestine include the following: absorbing water from the bolus (which is a round mass of organic matter passed down from the small intestine), storing feces in the rectum prior to excretion, and metabolizing undigested polysaccharides to short-chain fatty acids, which are passively absorbed for energy use. (3)
The large intestine is divided into three main parts: the cecum, the colon, and the rectum. The cecum, also known as the first part of the large intestine, is a pouch-shaped member that connects the colon to the ileum (which is the last part of the small intestine). The colon, which serves as a storage tube for solid wastes, is divided into four subcategories: the ascending colon, the transverse colon, the descending colon, and the sigmoid colon. The ascending colon, which is continuous with the cecum, extends upward towards the under surface of the liver. Then, the transverse colon, which is the longest part of the colon, passes downward near the lower end of the spleen. Next, the descending colon runs further down along the lateral border of the left kidney. When it reaches the lower end of the kidney, the colon turns toward the lateral border of the psoas muscle, where it will connect to the sigmoid colon. The sigmoid colon forms a loop of about 40 centimeters and lies within the pelvis region. Last but not least, the rectum. The rectum is the final straight portion of the large intestine that terminates in the anus. As mentioned before, this is where the feces are stored before being expelled out of the body. (1)
Moving on, the pH of the large intestine varies between 5.5 and 7.0, which indicates a fairly neutral environment. This is different from that of the small intestine, which exhibits a pH of 8.5, enabling absorption in mild alkaline environments; thus, water absorption in the large intestine occurs optimally around a neutral pH. (4)
In addition, the temperature inside the large intestine tends to be between 37-40°C. This is crucial to the breakdown of undigestible fibers, as hyperthermic or hypothermic temperatures proved to depress the catalysis of these carbohydrates. Therefore, the physical conditions in the large intestine are reasonably stable in order to ensure proper digestion of food. (4)
Who lives there?
Lactobacillus
Lactobacillus is a microbe that aids in production of lactic acid through homolactic fermentation, and can be found in the human gastrointestinal tract. This Gram-positive, rod shaped, bacterium is a beneficial bacterium that assists in enzymatic production to help with digestion, maintain pH balance and aid in healthy bacterium replacement in the gastrointestinal tract. The lactic acid produced by carbohydrate fermentation from lactobacillus, aids in maintaining the correct homeostatic pH to ensure no phage is able to survive at the low pH. Studies also show that it can synthesize necessary vitamins and help lower cholesterol levels (Ray). Lactobacillus also has been observed to degrade carcinogens, reduce or prevent carcinogenesis through antimutagenic activities. One of many strains of Lactobacilli that is commercially distributed is Lactobacillus Acidophilus, which is categorized as a probiotic. Probiotics are bacterium that can be taken as a supplement to help increase the beneficial bacteria that are normally present in their environment. In the case of Lactobacillus Acidophilus, it has fermentative capabilities, but lacks the ability to synthesize most cofactors and vitamins; which are expected normal capacities of microbes living in such a nutrient-rich niche such as the human gastrointestinal cavity (Altermann). The adherence of Lactobacillus Acidophilus to the cells lining the colon prevents binding of enteropathogenic and enterotoxigenic pathogens, promoting healthy metabolic activities. Lactobacillus Acidophilus and other strains of Lactobilli are widely used for treatment in diarrheal diseases, however the mixed results do not allow an exact conclusion on its treatment accuracy (Macfarlene). (5)
Bifidiobacterium
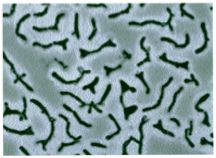
Bifidobacterium is also considered a probiotic bacterium that inhabits the anaerobic environment of the human gastrointestinal tract. The amount of this bacterium present decreases with age; thus, higher amounts are found in infants, and lower amounts in adults, which may lead to the needs of a probiotic supplement. Bifidobacterium is also a rod club-shaped (as pictured), Gram-positive cell, which has a symbiotic relationship with the host (gastrointestinal tract), benefiting both bacteria and host. This symbiotic relationship is seen by its adhesive ability to the microflora, the metabolism of undigested dietary carbohydrates and prevents pathogen colonization. This type of beneficial bacteria, similarly to the Lactobacilli, prevents binding of pathogens such as Escherichia coli, and Salmonella typhimurium, decreasing susceptibility to possible illnesses such as E. Coli poisoning in humans. The microflora normally lining the gut serves as a protective barrier against pathogens, however this can be compromised due to antibiotic treatments, stress, poor diet or other physiological distress. The Bifidobacteria can serve as resistance mechanism against the colonization of pathogens in the large intestine (Macfarlene). It has been observed that their competitive nature, against other gastrointestinal bacteria, is due to its ability to scavenge for a large variety of nutrients to use for energy (Schell). Studies also show that Bifidobacterium has also has potential to prevent cancer by reactivity with certain carcinogens and promotes immune stimulation (O'Sullivan). (6)
Methanogens and Sulfur reducing bacteria
In addition, methanogenic bacteria (e.g. Methanobrevibacter smithii) also benefit the gastrointestinal tract in that they reduce carbon dioxide and hydrogen gas to produce methane gas and water. This reduction reaction is an imperative characteristic of the large intestine as it aids in the fermentation of organic matter to obtain energy. For example, prior to entering the large intestine, the small intestine cannot digest or absorb dietary fibers for energy production; thus, these carbohydrates prove to be useless up to this point. However, once the indigestible sugars are in the large intestine, they will be fermented by gut organisms to break down the complex polymers (e.g. resistant starches, non-starch polysaccharides, oligosaccharides and etc.) into its monomeric constituents. These monomers are then oxidized to short chain fatty acids (SCFA), lactate, succinate, ethanol, hydrogen gas and carbon dioxide. The resulting SCFA can then enter central metabolic pathways where it can be converted into energy in the host organism. (7)
An important fact to note here is that hydrogen gas and carbon dioxide are products of the fermentation reaction. Since the large intestine is an anaerobic environment, the reducing energy is stored in the form of ethanol, lactate, succinate, or H2, but not in water (as would be the case in an aerobic environment). This poses a problem as accumulation of H2 inhibits oxidation of pyridine nucleotides, which leads to a redundant amount of substrate level phosphorylation. Therefore, in order to avoid this unnecessary energy expenditure, a balance between fermentation and H2 removal is imperative; and this where methanogens come in to save the day. (7)
Methanogens live symbiotically with the large intestine. The bacteria grow by reducing carbon dioxide and H2 to produce methane gas and water; hence, the fermentation products now function as nutrients for these organisms and not inhibitory poisons to the environment. The large intestine benefits from the reducing activity of the methanogens because methane is an effective pathway for H2 disposal, which relieves the inhibition of nucleotide oxidation. As a result, superfluous use of energy is avoided. (7)
An alternative to methanogenesis is the sulfate reducing pathway. The main substrates for sulfate reducing bacteria are also fermentation products; however, the product of this reduction pathway is a highly toxic hydrogen sulfide which can damage the colonic epithelium. The main organisms that perform this reaction include Desulfovibrio, Desulfobacter, Desulfomonas, Desulfobulbus, and Desulfotomaculum. (8)
Fermentation experiments in a lab revealed organisms competing for gaseous nutrients, as sulfate reducing bacteria overpowered methanogens when present in the same environment. However, recent experiments have shown methanogens displacing other H2 consuming bacteria in fecal slurries. In such experiments, the expression of sulfate reducing bacteria was probably limited or nonexistent because it is unlikely for methanogens to competitively overrule them. The reason for this is due to sulfur reducing bacteria having a higher affinity for H2 relative to that of methanogens. Therefore, determination of the more predominant organism depends on the sulfate concentration of the environment. The higher the sulfate concentration, the more reduction performed by sulfate reducing bacteria. (8)
Bacteroides
Bacteroides are anaerobic organisms that are characterized by the gram-negative cellular envelope. These bacteria exist symbiotically with the large intestine because the Bacteroides support the core metabolic activities (i.e. amino acid biosynthesis, carbon-oxygen lyases, membrane transport, and etc.) while feeding off dietary fibers passing through the colon. The common species of Bacteroides include B. vulgatus, B. distasoni, B. thetaiotaomicron, and B. fragilis. (9)
These organisms primarily feed off of plant polysaccharides and the host’s mucous glycans. (9) Fibrous polymers that pass through the large intestine can be only be broken down by Bacteroides. Once these sugars are in the large intestine, the Bacteriodes metabolize them into their respective monomers so that they can enter central metabolism for energy synthesis. (10) A unique trait of B. thetaiotaomicron is that it can use various polysaccharides to survive. The physiological explanation for this is that B. thetaiotaomicron possesses 226 glycoside hydrolases and 15 polysaccharide lyases. Note that B. thetaiotaomicron does not reside in human intestine; thus, we cannot digest such polysaccharides.
Bacteroides are also saccharolytic, meaning that they can obtain energy, carbon sources, dietary carbohydrate molecules from the host. (11) The products of fermentation reactions include acetate and propionate, which are short chains of fatty acids can then be absorbed via the large intestine. (11) B. distasonis are more specific for carbohydrate processing with the enzymes CAZy glycoside hydrolase family 13 and family 73.2 These enzymes aid in breaking down food molecules and supplying vitamins for metabolic purposes. (11) It also has more polysaccharide deacetylases, which allow it to deacetylate the O-acetylated sugars of the host’s epithelial glycans to utilize them.
Enterococcus
Common Species: E. avium, E. durans, E. faecalis, E. faecium, E. solitarius
Enterococci are gram-positive cocci that are found in many different places – normally found in the feces of people and many animals. Two major species – E. faecalis and E. faecium – can cause urinary tract infections and wound infections in human and animals most commonly. E. faecalis contributes about 90-95% of all species and E. faecium contributes about 5-10%. They can be also the cause of other diseases – bacteraemia (blood stream), endocarditis (heart vales), and meningitis (brain) – which occurs mostly in severely sick people with weak immune system. (12) Many of these Enterococcus infections have been commonly found in clinical environment. Healthy people with strong immune system will not be affected by Enterococci but they many become potential carriers.
Enterococci infections can be treated with various types of antibiotics, but unfortunately some of the Enterococci have become resistant to many types of antibiotics. They are resistant to β-lactam-based antibiotics (penicillin and cephalosporin) as well as many aminoglycosides. (13) In recent time, some of the Enterococci are found to be resistant to Vancomycin, and they are called VRE (Vancomycin-Resistant Enterococcus). VRE are especially dangerous because they can easily transmit the resistant genes from another (transformation or conjugation), so if there is small portion of Enterococci that are resistant to antibiotics, then the whole community will become resistant and no other antibiotics are effective. (14)
Enterococci are commensal bacteria inhabiting in the intestines of both humans and animals. Enterococci are capable of living in extremely hardy conditions that most of other bacteria won’t survive. They are likely to inhabit in the bowels of animlas or humans, and they are also found in soil, vegetation, and surface water (mainly due to contamination by animal feces). Enterococci are capable of growing at a range of temperatures from 10-45 °C and can grow in any environmental concentrations – hypotonic or hypertonic – and over broad range of pH – acidic or alkaline. Even though they are anaerobes, they can live in both low and high oxygen environments. In fact, Enterococci can survive at 60 °C for half an hour. They are also capable of living under high salt concentration (6.5% NaCl) and in high bile salts (40%). Some of them are naturally antibiotic resistant. (15)
Enterococci are anaerobes. They ferment carbohydrates to produce lactic acids (lactic acid bacteria). Enterococci living in large intestine will use undigested sugars from small intestine and indigestible fibers for their fermentation process. Especially for E. faecalis, it contains a large number of sugar uptake systems (PTS: phophoenolypyruvate phosphotransferase system) which recognize sugars outside and transport them with phosphorlyation. This process will use energy (ATP) more efficiently as it is compared to sugar transport by other non-PTS system. (16) E. faecalis can metabolize following types of sugars: D-glucose, D-fructose, lactose, maltose, etc. It also has cation homeostasis mechanism that contributes to its resistance to change in pH, salt concentration, etc. They under go fermentation process usually because they lack ability to process Kreb’s cycle and following electron transport chain reaction. (17)
Escherichia Coli
Common Species: Escherichia albertii; E. blattae; E. coli; E. fergusonii; E. hermannii; E. senegalensis; E. senegalensis; E. vulneris; E. sp.
Escherichia coli, also known as E. coli, is an anaerobic gram-negative bacterium that can be found in large intestine of warm-blooded animals and humans. Even though it is a predominant and consistent organism, it contributes to only small portion of the content of total bacteria in GI tract. It is often used as an indicator to test the fecal contamination in nature such as water since E. coli is able to survive outside the body for a brief time period.(18)(19) Because of its simple nutritional requirement and ability to grow rapidly, E. coli is also useful in studying the organisms’ essential processes of life.(20)
E. coli are both pathogenic and nonpathogenic. Some are nonpathogenic, but it might cause infection if the bacterium is introduced to other tissues in a debilitated host; however, the bacteria from contaminated water or undercooked meat may cause infection in a healthy person as well. (21) Some virulent E. coli can even lead to urinary tract infections, neonatal meningitis, diarrheal disease, and gastroenteritis. (22) They damage the host in several steps: colonizing the intestinal mucosal surface, evading the defenses by the host, and lastly multiplying themselves in numbers.(21) Since the bacterial infections are usually treated with antibiotics, the antibiotic-resistance became problematic. E. coli often stay with diverse species of bacteria in a form of biofilm and this causes the transfer of antibiotic-resistant plasmids of E. coli to other bacteria such as Staphylococcus aureus. (23)
Despite the fact that E. coli is a unicellular organism, it has an amazing ability in interacting with the environment. It can respond to such signals including pH, osmolarity, temperature, and chemicals. With its chemotaxis proteins in cytoplasmic membrane, E. coli can detect attractants and repellants without any stimuli. With around six flagella, it can swim towards the attractant with counterclockwise flagellar rotation, but bacteria tumbles with clockwise rotation when it senses the repellant.(24) It can also adjust the size of the porins on the outer membrane in order to regulate the temperature and osmolarity by importing or exporting the larger substances such as nutrients. E. coli can detect the chemicals in the surrounding environment with its complex metabolism mechanism.(24)
E. coli is able to survive with or without oxygen. Unlike aerobic organisms which use oxygen as the final electron acceptor, E. coli use nitrate or other molecules. The major determinants of organization of bacterium are the type of terminal electron acceptor and the presence of glucose (Barrett). E. coli demonstrates only six distinct functional states no matter what carbon source or electron acceptor has been used. (23) Unless the carbon sources are available, they do not wastefully make the enzymes for degradation. If the metabolites are available in the environment as nutrients, they also don’t make the enzymes for synthesis. (18)
Clostridium
Common Species: Clostridium dfficile, Clostridium tetani, Clostridium perfringens, Clostridium acetobutylicum
Clostridium, is a gram-positive bacteria belonging to the family of Clostridiaceae. It is capable of forming endospores during extreme environmental conditions. These organisms are capable of surviving in many environments, but are predominantly found in soil and feces; be that as it may, Clostridium prefer to live in the human body as such an environment provides optimum growth for the organism. (25)
Most of the species in Clostridium are pathogenic. Clostridium deficile for instance, may cause inflammation in the colon of the gastrointestinal tract. Ironically, treatment with relevant antibiotics does not kill this organism, but speed its the growth. The only factor that limits its growth is the presence of other bacteria in the colon (i.e. other competitors). (25)
Clostridium is able to survive without oxygen. Unlike aerobic organisms which use oxygen as the final electron acceptor, Clostridium use nitrate or other molecules. These organisms are usually pathogens that live freely in our gastrointestinal tract. A trademark characteristic of this organism is that it is the major cause of Pseudomembranous Coliits, an inflammatory disease of the colon characterized by exudative plaques on congested mucosa. (25)
Studies have shown that commercial broiler chickens that were vaccinated with Clostridium perfiringens type A Toxoid controlled the growth of necrotic enteritis over a 65 week period. Sera and egg yolk that were injected with the vaccine had higher antibody for C. perfringers compared to controls.
What other organisms are present (e.g. plants, fungi, etc.)
Fungi
Candida species, such as C. Albicans, C. Glarata, are one of the few fungal organisms that live in the large intestines. Candidiasis is a condition characterized by yeast/fungal overgrowth that can result in symptoms such as incapacitating fatigue, problems with concentration, extreme tightness in shoulders and neck. Candida spicies are similar to brewers yeast having dimorphic stages (grow in two morphological forms), allowing them to become a yeast cell form or in a hyphae form. Changes in enviromental conditions such as pH, temperature, or compound availability changes, the organisim can switch in morphological form. If the pH of the environment is about 4.5, the organism will be budding in the yeast form. If the pH is raised to 6.7, the Candida would switch gene expression and begin growing in the hyphae mode. The ability to switch gene expressions ensures the organisms pathogenicity. (26)
Because Candida reside in mucosal communities, they live next to the host tissue and interact with the gut immune system. An important fact to note is that many intestinal organisms have immunomodulatory properties, which allow them to affect the activities of the intestinal epithelial cells (B and T lymphocytes) as well as other accessory cells from the mucosal immune system. (27)
Despite Candida’s extraordinary ability to evade cell death, its growth can be maintained with the presence of bifidobacterias and similar probiotics. The probiotic bacteria interact with Candida by preventing the organism to propagate uncontrollably. This in turn will reduce inflammatory responses of the intestinal tracts and relieve some of the common symptoms. When Candida proliferate exponentially, this is an implication that there is a deprived presence of bifidobacterias and other probiotics. (28)
Candida, like most large intestine residents, are anaerobes due to the oxygen lacking environment. Therefore, their primary metabolism is that of anaerobic metabolism. An interesting fact to note is that Candida are similar to yeast, in that they utilize fermentation.
Protozoa
Protozoa are a collection of single celled eukaryotic organisms. They are among the simplest of all living organism. Most protozoans reside in soils and aquatic habitats. Protozoa are usually not present in normal human intestine but some are capable of colonizing the intestine as parasites. The organisms that reside in the intestine predominantly affect individuals living in the tropics. These are thought to cause diarrhea and other gastrointestinal issues.
Entamoeba histolytica is a class of protozoan that usually resides asymptomatically in the large intestine; however, when it penetrates the intestinal mucosa causes an invasive intestinal or extraintestinal infection called amebiasis. In 1993, E. histolytica was reclassified into two distinct species based on epidemiological and biochemical data: E. histolytica (pathogenic) and E. dispar (nonpathogenic). It is now believed that the overall prevalence of infection is 90% of E. dispar and 10% of E. histolytica.
“Entamoeba parasites have a two-stage life cycle consisting of a disease-causing trophozoite stage and an infectious cyst stage.” Nataro (34) The organism enters the gastrointestinal tract via ingestion from fecal contaminated food and water. The E. histolytica ingested in a cyst stage is resistant to gastric acidity, chlorination, and desiccation. In the intestinal lumen, the organism escapes from the cyst to produce trophozoites by asexual reproduction. Then the trophozits adhre to the the intestine's thick and dense mucous layer.
Entamoeba may enclose back to a cyst in the mucin layer so that the cysts can be excreted in the stool. Living the stool will ensure the life cycle of the Entamoeba if it is ingested by another oganism. E. histolytica manages to penetrate the mucin layer and contact “colonic epithelial cells via a galactose- and N-acetyl-D-galactosamine (Gal/GalNac)-specific lectin.” Nataro (34) The adhered trophozoites cause epithelial cell death due to the secretion of virulent factors such as amoebapores and cysteine proteinase.
An important fact to note is that E. histolytica rarely enter the portal circulation because in the large intesetine, it interacts with bacterial flora, which inhibits the invasion of E. histolytica by decreasing luminal proteases. Thus, the E. histolytica will not be able to adhere to the mucin layer as anticipated. Wiesner (35) Presumably, alterations in bowel flora results in outbreak of amebiasis, implying that the protective role of bacteria needs further study.
Current Research
Probiotics for Irritable Bowel Syndrome: The affects of intestinal bacteria on colonic health
As discussed earlier, there are a myriad of bacterium that reside in the human colon and are carried out through various mechanisms to contribute to digestion. Lactobacilli and Bifidobacterium are two types of bacteria that benefit the health of the host in a symbiotic relationships with the gut flora lining the colon. These beneficial bacteria have been researched and observed in order to utilize their health benefits on diseased or irritable colons. Diet will always play a factor in maintaining colonic microflora health, however, this may not be a solution for those who suffer from chronic colonic digestion issues.
Commercial probiotics have been one of the many pharmaceutical interventions that have been studied to possibly promote better colonic health. These commercially produced probiotics are typically prepared with a mixture of lactobacilli and bifidobacteria, which are found to aid in break down of carbohydrates to directly interact with the host metabolism. Research has been ongoing in observing the relationship between the immune system and the intestinal microbes in hopes of finding an immune system-bacterium symbiotic relationship. Typically the host's immune system (a human's in this case) has the ability to differentiate between normal bacteria and pathogenic bacteria via specific receptors. Toll-like receptors (TLRs) have different types of receptors found on Gram-positive and Gram-negative cells to mediate recognition responses on bacterial DNA sequences. Through this method, the host's immune system holds the initiates protection from infections when pathogens are present. Be that as it may, when the probiotic bacterium is recognized it was observed to have an anti-inflammatory effect on the intestinal mucosal surface through various mechanisms. Studies show that the probiotics have the ability to alter the gut flora by increasing the secretion of intestinal mucus and by changing the volume composition of the fermented stool and gas fermented. (29)
Irritable Bowel Syndrome (IBS) is only one of the many colonic irritations that can occur in the human gastrointestinal tract, and it has been hypothesized that probiotics are able to benefit suffering patients. The probiotics also have an antiviral and antibacterial effect, which could potentially benefit the 15-25% of patients who suffer from infectious gastroenteritis. As mentioned above, the change in stool composition and alteration in gut flora activity could aid with IBS treatment. (30)
These theoretical ideas of the probiotics benefitting those suffering from IBS by these mechanisms were tested through various clinical trials in order to understand the actual target. Using a composite probiotic (VSL#3), the first study focused on diarrhea-predominant IBS patients who received placebos, or received the VSL#3 powder twice daily over an eight week span. Of the 25 patients in this study, no differences were observed in gastrointestinal transit time, bowel function scores, or overall relief of original symptoms. However, there was a difference observed on abdominal bloating in the placebo versus active group. The second trial included 48 patients, again using the active treatment or placebos for 8 weeks. There was an increase in the number of patients who experienced relief in flatulence, but lower numbers were recorded in relief of symptoms of abdominal pain. 46% of the patients in the active treatment and 33% of the placebo patients reported satisfactory relief for half of the weeks. (30)
A different research group used 362 women with any subtype of IBS to receive one of three doses of B. Infantis or placebo once daily for 4 weeks. The results showed that in the last week, only one of the active subgroups had reported significant improvements in comparison to baseline standards. Some of these improvements included relief of abdominal pain/discomfort, bloating/distention, and bowel movement difficulty. (30)
To study the role of inflammation in probiotics another study was performed focusing on peripheral blood cytokine levels; 75 patients were given B. Infantis or L. Salivarius or placebo for 8 weeks. The Bifidobacterium showed superior symptom relief to that of the Lactobacillus active group; these symptoms include the three dominant symptoms associated with IBS: Bloating/distention, abdominal pain/discomfort and bowel movement difficulty. Levels of interleukin (IL)-10 and IL-12 were measured to observe the inflammatory response. The ratio of lower IL-10 and increased IL-12 levels to that of the baseline control, demonstrated a pro-inflammatory cytokine profile. It was reported that the bifidobacteria treated group had restored a normal IL-10 / IL-12 ratio, but the placebo and L. salivarius treated group had a persistent pro-inflammatory cytokine profile. This observation was summarized by the authors in saying, "by demonstrating a normalization of the IL-10 / IL-12 ratio in the bifidobacteria-fed subjects alone, and in parallel with symptomatic improvement, we provide the first evidence for efficacy for an anti-inflammatory approach in IBS" (Walsh). (30)
Microbially Triggered Drug Delivery to the Colon
Oral therapy has been the most popular route for drug delivery. However, colon, the terminal part of the GI track has been recognized a new site for drug delivery including novel therapeutic drugs. Over 400 bacterial species inhabit in the gastrointestinal (GI) tract to have specific niche throughout the tract. Colon is a part of intestine where a large group of gram negative microflora is found. The flora produces numerous enzymes that are excreted for formulation of colon-specific drug delivery systems. This article focused on analyzing various microbially activated drug delivery systems for colon specific drug delivery with specific reference to the microflora of the GI track and their role of targeting drug delivery to the colon. (31)
Since the anaerobic bacteria of the colon are capable of reacting with complex carbohydrates entering the colon, many different systems have been established for drug delivery to the colon. So the prodrugs are tested on different systems (the carriers) that are based upon biodegradable polymers and polysaccharides. In conclusion, the biodegradable polymers (azo and other synthetic polymers) need detailed toxicological study to be performed before they are commercially used. Polysaccharides without any toxic properties are an alternate way of coating drugs (carriers for prodrugs). However, polysaccharide with moieties can bind to specific regions (the inflamed colonic lesions or cancer tissues) before it is being effective. (31)
Increase of Intestinal Bifidobacterium and Suppression of Coliform Bacteria with Short-Term Yogurt Ingestion
A known carcinogen is Coliform bacilli, which causes build-up of bile acid degradation products. This accumulation of undesired products may lead to colon cancer. Certain anaerobic bacteria are beneficial to humans because they produce short-chain fatty acid (via fermentation of indigestible carbs), which lowers the intestinal pH and suppresses the colonization of harmful bacteria like Coliform bacilli. Yogurt contains large amounts of lactic acid bacteria (107 per milliliter to be precise), however, not all bacteria from yogurt can survive the extreme acidic condition of the stomach. Bacteria like Lactobacillaceae and streptococcaceae will most surely die in the presence of gastric acid and will not propagate in favorable numbers. Bifidobacteria, on the other hand, have proved to be survivors of the harsh conditions of the stomach and may continue to thrive in the large intestine. (32)
The research question concerning the article at hand deals with whether we can increase the amount of Bifidobacteria via injection of yogurt, and also if an increased amount of Bifidobacteria in the large intestine decreases the Coliform bacilli proliferation. This experiment was performed on 34 healthy people (4 males and 30 females from ages 19 to 42) in a span of 26 days. Subjects were asked to consume AB yogurt containing equal amounts of Streptococcus thermophilus, Lactobacillus bulgaricus, Lactobacillus acidophilus, and Bifidobacterium bifidum for the middle 10 days, while ingesting no yogurt for the first and last 8 days. Their stool samples were collected in 3 to 4-day intervals to test for the presence of any bacteria. The purpose of collecting the stool samples was to verify which bacteria were able to get through the stomach and pass through the large intestine. The result was that only Bifidobacterium bifidum were identified to be in all the stools starting 3 days after the initial yogurt ingestion and until after 8 days of no yogurt ingestion. Also, during the 10 days of yogurt ingestion, the amount of anaerobic bacteria in the stool increased significantly, while a halt in yogurt consumption caused the concentration of anaerobes to return to normal levels. In contrast, the amount of aerobic and Coliform bacteria decreased during the yogurt ingestion period, but returned back after stopping of yogurt ingestion. (32)
Researchers concluded that Bifidobacteria are able to survive the stomach’s low pH levels, which allow the organism to proliferate in the large intestine. Bifidiobacteria also have anti-cancer effect as it suppresses the colonization of the aerobe, Coliform. Amount of Bifidobacteria in large intestine can be increased by ingestion of yogurt containing such bacteria. (32)
Ability of Lactobacillus fermentum to overcome host α-galactosidase deficiency, as evidenced by reduction of hydrogen excretion in rats consuming soya α-galacto-oligosaccharides
Soya contains high concentration of α-galacto-oligosaccharides but human and other mammals lack α-galactosidase to digest the oligosaccharide in the small intestine. As these carbohydrates enter the large intestine, α-galactosidase-producing lactobacilli can use them for fermentation. This article was “to assess the ability of α-galactosidase-producing lactobacilli to improve the digestibility of α-galacto-oligosaccharides” by setting up experimental systems on rats. (33)
To test their hypothesis, a model experimental system to assess the potential of α-galactosidase-producing lactobacilli to improve α-galacto-oligosaccharides digestion in rats was set up. The rats were fed with soy milk orally and placed in respiratory chambers to monitor their fermentative gas excretion. Gnotobiotic rats with a hydrogen gas producing strain (Clostridium butyricum) were first checked for their validity in this experiment. Significant amount of hydrogen (H2) emission was detected after ingestion of native soy milk, but no significant amount of H2 emission was detected after ingestion of α-galacto-oligosaccharide-free soy milk. This step validated the experimental system. The rats did not produce H2 as the native soy milk was fermented using the α-galactosidase-producing Lactobacillus fermentum CRL722 strain. This also validated the bacteria used in the experiment. A significant reduction of H2 emission was found when L. fermentum CRL722 was coadministered with native soy milk (reduced by 50%). This also explained that α-galactosidase from L. fermentum CRL722 remained active in the gastrointestinal tract of rats associated with C. butyricum. L. fermentum CRL722 induced a significant reduction in H2 emission by 70% in human-microbiota associated rats. (33)
In conclusion, the results suggest that L. fermentum α-galactosidase is able to relieve α-galactosidase deficiency in rats very partially. That means that this can be used in further applications where “lactic acid bacteria could be used as a vector for delivery of digestive enzymes in man and animals.” (33)
Summary
Due to the oxygen lacking characteristic of the large intestine, the organisms that thrive in this niche are anaerobes. At first blush, these organisms appear to be like any other microorganism that propagate in extreme conditions; however, upon closer inspection, these organisms are exemplaries of adaptation. This is due to their ability to not only withstand the gastric acids of the stomach, but the subsequent alkaline pH of the small intestine followed by the neutral pH of the large intestine. Therefore, the large intestine is home to no oridnary microbes.
The microbes mentioned thus far proved to be either beneficial (e.g. Methanogens and Sulfur reducing bacteria), pathogenic (e.g. Candida) or both (e.g. E. coli). The pathogenic oraganisms of the large intestine tend to have irreversible health effects to those who are infected. Thus, it may sound appealing to insulate ourselves from all bacteria; but, there are also microbes that play an imperative role in digestion, so we cannot live without them.
References
1. Medline Plus: "Large Intestine": http://www.becomehealthynow.com/article/bodydigestive/787/
2. About: Colon Cancer: "Large Intestine": http://coloncancer.about.com/od/glossaries/g/Large_Intestine.htm
3. Wikipedia: "Large Intestine": http://www.becomehealthynow.com/article/bodydigestive/787/
4. Wikipedia: "Colon (anatomy)": http://en.wikipedia.org/wiki/Colon_(anatomy)
5. Altermann, Eric. "Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM.": http://www.pnas.org/content/102/11/3906.full
6. Schell, MA. "The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract" Proc Natl Acad Sci U S A. 2002 Oct 29;99(22):14422-7. Epub 2002 Oct 15: http://www.ncbi.nlm.nih.gov/sites/entrez
7. Gibson, G.R.;Macfarlane, G.T.; Cummings, J.H. "Sulphate reducing bacteria and hydrogen metabolism in the human large intestine.": http://www.pubmedcentral.nih.gov/pagerender.fcgi?artid=1374298&pageindex=1
8. Gibson, G.R.;Macfarlane, G.T.; Cummings, J.H. "Sulphate reducing bacteria and hydrogen metabolism in the human large intestine.": http://www.pubmedcentral.nih.gov/pagerender.fcgi?artid=1374298&pageindex=2#page
9. Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, et al. “Evolution of Symbiotic Bacteria in the Distal Human Intestine.” PLoS Biology Vol. 5, No. 7 (2007)
10. Stafferton, Joanne. "Polysacchardies: Their Structure and Function": http://molecular-biology.suite101.com/article.cfm/polysaccharides
11. East Carolina University: "What are Bacteroides?": http://borg.med.ecu.edu/~webpage/about.html#Bacteroides
12. Medterms: "Definition of Enterococcus": http://www.medterms.com/script/main/art.asp?articlekey=20162
13. Ryan KJ, Ray CG (editors). 2004. Sherris Medical Microbiology, 4th ed., McGraw Hill, 294-295.
14. Ronnie Falcao, Sept. 1997. "VRE – Vancomycin-Resistant Enterococcus": http://www.gentlebirth.org/vre/vremain.html
15. University of Oklahoma Health Sciences Center: "Pathogenicity of Enterococci": http://www.enterococcus.ouhsc.edu/lynn_revirew.asp
16. Michael Gilmore. 2002. The Enterococci: Pathogenesis, Molecular Biology, and Antibiotic Resistance. American Society for Microbiology Press.
17. De la Maza, Luis M., Marie T. Pezzlo, and Janet T. Shigei. 2004. Color Atlas of Medical Bacteriology. American Society for Microbiology Press.
18. Todar, K. "Pathogenic E. coli". Online Textbook of Bacteriology. University of Wisconsin-Madison Department of Bacteriology. Nov 2007.
19. "Escherichia coli O157:H7". CDC Division of Bacterial and Mycotic Diseases. Jan 2007.
20. Microbes in Norwich: Escherichia coli.
21. Nataro, James P. and James B. Kaper. 1998. "Diarrheagenic Escherichia coli." Clinical Microbiology Reviews, vol. 11, no. 1. American Society for Microbiology. (142-201)
22. Barrett, Christian L. and Herring, Christopher D. and Reed, Jennifer L. and Palsson, Bernhard O. “The global transcriptional regulatory network for metabolism in Escherichia coli exhibits few dominant functional states” University of California, San Diego Department of Bioengineering. Nov 2005.
23. Salyers AA, Gupta A, Wang Y (2004). "Human intestinal bacteria as reservoirs for antibiotic resistance genes". Trends Microbiol.
24. Maki, Nazli. “Motility and Chemotaxis of Filamentous Cells of Escherichia coli”. University of Wisconsin-Madison Department of Biochemistry, Chemistry, and Genetics. Jan 2000: http://jb.asm.org/cgi/reprint/182/15/4337.pdf
25. C.P. Tamboli, C. Neut and P. Desreumaux et al., Dysbiosis in inflammatory bowel disease, Gut 53 (2004), pp. 1–4.
26. R. Soll, David. The regulation of cellular differentiation in the dimorphic yeast Candida albicans. BioEssays. Volume5, Issue1, Date:1986, Pages:5-11.
27. Pla, J.; Gil, C.; Monteoliva, L.; Navarro-Garcia, F.; Sanchez, M.; Nombela, C.; Understanding Candida albicans at the Molecular Level Yeast; Volume12, Issue16, Date:December 1996, Pages:1677-1702
28. Panagoda, G. J.; Ellepola, A. N. B.; Samaranayake, L. P.; Adhesion of Candida parapsilosis to epithelial and acrylic surfaces correlates with cell surface hydrophobicity Mycoses; Volume44, Issue1-2, Date:March 2001, Pages:29-35
29. Macfarlene, George T., & John H. Cummings "Probiotics & Prebiotics: Can Regulating the activities of Intestinal Bacteria Benefit Health?": http://www.pubmedcentral.nih.gov/tocrender.fcgi?iid=124807
30. Walsh, Nancy. "Probiotics for Irritable Bowel Syndrome.(ALTERNATIVE MEDICINE: AN EVIDENCE-BASED APPROACH).": http://find.galegroup.com/itx/start.do?prodId=EAIM
31. Sinha, V.R.; Kumria, Rachna; “Microbially Triggered Drug Delivery to the Colon.” European Journal of Pharmaceutical Sciences. Volume 18, Issue 1. January 2003. pg 3-18.
32. Chen R. M., et al. “Increase of Intestinal Bifidobacterium and Suppression of Coliform Bacteria with Short-Term Yogurt Ingestion.” Journal of Dairy Science 18 (1999): 2308-2314
33. LeBlanc, Jean Guy; Ledue-Clier, Florence; Bensaada, Martine, et al. "Increase of Intestinal Bifidobacterium and Suppression of Coliform Bacteria with Short-Term Yogurt Ingestion" BioMed Central Ltd. Vol. 8 Article no. 22. January 29, 2008: http://www.biomedcentral.com/1471-2180/8/22
34. P. Nataro, James; S. Cohen, Paul; L.T. Mobley, Harry: "Colonization of Mucosal Surfaces": http://books.google.com/books?id=a1w1pMa8eksC&pg=PA61&lpg=PA61&dq=Nataro,+James+P.,+and+Cohen+Paul+S.,+Mobley+Harry+L.+T.,+and+Weiser+Jeffrey+N.+Colonization+of+Mucosal+Surfaces.+Washington,+DC:+ASM+Press,+2005&source=web&ots=jjdPAXNV20&sig=7Zmme-4C07ar9FtV7PU0msKyJxw&hl=en&sa=X&oi=book_result&resnum=1&ct=result
35. Wiesner J.; Reichenberg A.; Heinrich S.; Schlitzer M.; Jomaa H.: "The plastid-like organelle of apicomplexan parasites as drug target.": http://www.ncbi.nlm.nih.gov/pubmed/18473835
Edited by Benjamin Dae Lee, Hilary Otorowski, Julia Son, Rebecca Son, Alexander Pang, Ella Chen, David Araiza, students of Rachel Larsen

