Microbial Colonization of Space Stations: Difference between revisions
No edit summary |
|||
| (17 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | |||
==Introduction== | ==Introduction== | ||
[[File:ISS.jpg|300px|thumb|right|International Space Station[[#References|[3]]]. Credit: NASA]] | [[File:ISS.jpg|300px|thumb|right|International Space Station[[#References|[3]]]. Credit: NASA]] | ||
Long-term [http://en.wikipedia.org/wiki/Space_station space station] missions to date include [http://en.wikipedia.org/wiki/Salyut_program Salyut], [http://en.wikipedia.org/wiki/Almaz Almaz], [http://en.wikipedia.org/wiki/Skylab Skylab], [http://en.wikipedia.org/wiki/Mir Mir], [http://en.wikipedia.org/wiki/International_Space_Station ISS] (International Space Station), and [http://en.wikipedia.org/wiki/Tiangong Tiangong], with the latter two still ongoing. | Long-term [http://en.wikipedia.org/wiki/Space_station space station] missions to date include [http://en.wikipedia.org/wiki/Salyut_program Salyut], [http://en.wikipedia.org/wiki/Almaz Almaz], [http://en.wikipedia.org/wiki/Skylab Skylab], [http://en.wikipedia.org/wiki/Mir Mir], [http://en.wikipedia.org/wiki/International_Space_Station ISS] (International Space Station), and [http://en.wikipedia.org/wiki/Tiangong Tiangong], with the latter two still ongoing. Researches on the microbial communities which persist during these missions have demonstrated that space stations support a diverse microbial community. While most microbes are not harmful, routine microbial monitoring onboard space stations is still essential in preventing material biodeterioration[[#References|[2]]] and maintaining crew health[[#References|[3]]]. As currently identified, these microorganisms originate from Earth, with the main source being from crewmembers themselves and from materials manufactured on ground.[[#References|[4]]] Environmental monitoring is important in identifying these microorganisms and providing data for future studies in attempt to minimize microbial threat to crewmembers’ performance and spacecrafts’ durability. | ||
==Physical Environment== | ==Physical Environment== | ||
[[File:Pathogen human biotope.png| | [[File:Pathogen human biotope.png|350px|thumb|left|Pathogen infestation on crewmembers. | ||
Higher than average indicates that for the given locational microflora, there is a higher percentage of microorganisms that are pathogens when compared to the percentage of pathogens in all microorganisms present on all habitats onboard. | Higher than average indicates that for the given locational microflora, there is a higher percentage of microorganisms that are pathogens when compared to the percentage of pathogens in all microorganisms present on all habitats onboard. | ||
| Line 11: | Line 19: | ||
Microorganisms exist in the portable water, ventilating air, and on surfaces of space stations. [[#References|[4]]] While these microorganisms share the same living space as the crewmembers, the crewmembers themselves also serve as incubators for various microbes. [[#References|[5]]] Researchers have identified several strains of bacteria and fungi as opportunistic pathogen that may pose threats to crewmember. (refer to [[#Key | Microorganisms exist in the portable water, ventilating air, and on surfaces of space stations.[[#References|[4]]] While these microorganisms share the same living space as the crewmembers, the crewmembers themselves also serve as incubators for various microbes.[[#References|[5]]] Researchers have identified several strains of bacteria and fungi as opportunistic pathogen that may pose threats to crewmember. (refer to [[#Key Microorganisms|Key Microorganisms]] section) Moreover, they have also identified the type of microbes responsible for biodegradation of structural materials.[[#References|[6]]] (refer to [[#Biodegradation|Biodegradation]] section) | ||
==Microbial Diversity and Interactions== | ==Microbial Diversity and Interactions== | ||
[[File:Biofilm formaiton.jpg |300px|thumb|right| Biofilm development associated with fungi and bacteria.]] | |||
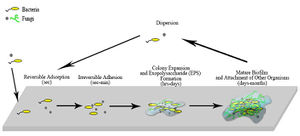
Other than in portable water and air, bacteria and fungi typically co-exist in [http://en.wikipedia.org/wiki/Biofilm biofilms] on surfaces, including water pipe lines.[[#References|[7]]] Biofilms provides suitable growth conditions, and intrinsically contribute to [http://www.nlm.nih.gov/medlineplus/antibiotics.html antibiotic] and [http://en.wikipedia.org/wiki/Biocide biocide] resistance[[#References|[13]]] due to horizontal gene transfer of antibiotic resistance plasmid, and the fact that microbes positioned inside biofilms are harder to reach. One study shows that as much as 36 species of fungi and 58 species of bacteria can co-exist in a single biofilm.[[#References|[16]]] However, the composition varies drastically due to dynamic (changing) environmental conditions, available nutrient (surface materials), and also the thickness of biofilm.[[#References|[16]]] In addition, exchange of microflora carried by crewmembers travelling to and from the space craft also increases microbial diversity onboard space stations.[[#References|[12]]] | |||
==Space Microbial Processes== | ==Space Microbial Processes== | ||
===Biodegradation=== | ===Biodegradation=== | ||
Biodegradation of the surfaces of spacecraft structural materials are results of colonization of bacteria and fungi. [[#References|[2]]] While fungi are more abundant in early colonization and biofilm formation on [http://en.wikipedia.org/wiki/Polymer polymeric materials], given sufficient humidity, subsequent microbes thrive in biofilms that forms at wide ranges of temperatures.[[#References|[2]]] Biodegradation of bacteria and fungi predominantly occur on these surfaces due to the availability of carbon in surface finishings, as well as other nutrients present in structural materials[[#References|[2]]] (polymeric-base and metal-based). In thin biofilms, oxic condition support aerobic biodegraders; in thicker biofilms, where oxygen diffusion is limited, anaerobic fermenters and metal reducers are come into play. Although these biodegradation of carbon (polymeric)-based and metal-based materials are due to similar hydrolysis reactions, the yielding products and degree of degradation are different depending on different microbes. [[#References|[2]]] | Biodegradation of the surfaces of spacecraft structural materials are results of colonization of bacteria and fungi.[[#References|[2]]] While fungi are more abundant in early colonization and biofilm formation on [http://en.wikipedia.org/wiki/Polymer polymeric materials], given sufficient humidity, subsequent microbes thrive in biofilms that forms at wide ranges of temperatures.[[#References|[2]]] Biodegradation of bacteria and fungi predominantly occur on these surfaces due to the availability of carbon in surface finishings, as well as other nutrients present in structural materials[[#References|[2]]] (polymeric-base and metal-based). In thin biofilms, oxic condition support aerobic biodegraders; in thicker biofilms, where oxygen diffusion is limited, anaerobic fermenters and metal reducers are come into play. Although these biodegradation of carbon (polymeric)-based and metal-based materials are due to similar hydrolysis reactions, the yielding products and degree of degradation are different depending on different microbes.[[#References|[2]]] | ||
===Antibiotics Sensitivity=== | ===Antibiotics Sensitivity=== | ||
[[File:Space Antibi.png]] | |||
According to various in vitro studies, the microgravitaional environment in space enhances microbial growth.[[#References|[1]]][[#References|[12]]] Several factors were examined in a closed, isolated environment on ground-based labs, and the results show a decreased lag phase and increased final yield compared to normal-gravity conditions.[[#References|[12]]] This enhanced microbial growth also contribute to increase of antibiotic resistance.[[#References|[1]]][[#References|[12]]] In addition to the confined, microgravitaional condition, various unknown factors onboard also provide distinctive selective pressures that favor certain microbes that eventually become harder to eliminate.[[#References|[12]]] | |||
==Key Microorganisms== | ==Key Microorganisms== | ||
===Microbe Identification Techniques (Onboard Environmental Monitering)=== | ===Microbe Identification Techniques (Onboard Environmental Monitering)=== | ||
[[File:Locad-device.jpg|200px|thumb|right|LOCAD‐PTS Hardware: Reader and Cartridge[[#References|[2]]]. Credit: NASA]] | |||
More recently, ISS launched [http://ntrs.nasa.gov/search.jsp?R=20090017684 Lab-On-A-Chip Application Development Portable Test System] (LOCAD-PTS). [[#References|[11]]] This new technology can be hand-held and allowed rapid microbial monitoring of bacteria and fungi since its implementation in December 2006. [[#References|[11]]] | Most antimicrobial and environmental monitoring implementations on ISS are results of studies done by previous expedition, Mir and Skylab. Before December 2006, isolation of microorganisms depended on the growth of environmental isolates on agar media onboard space stations and the subsequent ground-based analysis back on Earth.[[#References|[11]]] However, this method restricts the number identifiable microorganisms since more than 95% of environmental microbes do not grow on conventional agar growth media. Furthermore, accuracy is limited since months often passed before returning to Earth for further analysis.[[#References|[11]]] | ||
More recently, ISS launched [http://ntrs.nasa.gov/search.jsp?R=20090017684 Lab-On-A-Chip Application Development Portable Test System] (LOCAD-PTS).[[#References|[11]]] This new technology can be hand-held and allowed rapid microbial monitoring of bacteria and fungi since its implementation in December 2006.[[#References|[11]]] | |||
===Portable Water=== | ===Portable Water=== | ||
Dominant bacteria are [http://hamap.expasy.org/proteomes/ACISJ.html Acidovorax spp.], [http://patricbrc.org/portal/portal/patric/Taxon?cType=taxon&cId=1033 Afipia spp.], [http://patricbrc.org/portal/portal/patric/Taxon?cType=taxon&cId=41275 Brevundimonas spp.], [http://ijs.sgmjournals.org/content/59/11/2778.full Propionibacterium spp.], [http://www.phac-aspc.gc.ca/lab-bio/res/psds-ftss/serratia-spp-eng.php Serratia spp.], [[#References|[14]]] [[Sphingomonas spp: Agents of Bioremediation and Pathogenesis|Sphingomonas spp.]], and [[Methylobacterium|Methylobacterium spp.]], [[#References|[5]]] and opportunistic human pathogens include Afipia spp., Brevundimonas spp., Propionibacterium spp., Serratia spp., and Sphingomonas spp. (rarely pathogenic to human). | Dominant bacteria are [http://hamap.expasy.org/proteomes/ACISJ.html Acidovorax spp.], [http://patricbrc.org/portal/portal/patric/Taxon?cType=taxon&cId=1033 Afipia spp.], [http://patricbrc.org/portal/portal/patric/Taxon?cType=taxon&cId=41275 Brevundimonas spp.], [http://ijs.sgmjournals.org/content/59/11/2778.full Propionibacterium spp.], [http://www.phac-aspc.gc.ca/lab-bio/res/psds-ftss/serratia-spp-eng.php Serratia spp.],[[#References|[14]]] [[Sphingomonas spp: Agents of Bioremediation and Pathogenesis|Sphingomonas spp.]], and [[Methylobacterium|Methylobacterium spp.]],[[#References|[5]]] and opportunistic human pathogens include Afipia spp., Brevundimonas spp., Propionibacterium spp., Serratia spp., and Sphingomonas spp. (rarely pathogenic to human). | ||
===Air=== | ===Air=== | ||
| Line 40: | Line 71: | ||
===Surfaces=== | ===Surfaces=== | ||
Dominant bacteria are Staphylococcus spp., [[#References|[5]]] [http://en.wikipedia.org/wiki/Pseudomonas_aeruginosa Pseudomonas aeruginosa], [[Ochrobactrum anthropi|Ochrobactrum anthropi]], [[Alcaligenes|Alcaligenes denitrificans]], [http://medical-dictionary.thefreedictionary.com/Xanthomonas+maltophilia Xanthomonas maltophila], and [http://en.wikipedia.org/wiki/Vibrio_harveyi Vibrio harveyi][[#References|[2]]], while dominant fungi are Aspergillus spp. And [http://en.wikipedia.org/wiki/Cladosporium Cladosporium spp.][[#References|[5]]] Opportunistic human pathogens included Staphylococcus spp., Pseudomonas aeruginosa, Ochrobactrum anthropi, Alcaligenes denitrificans (rarely pathogenic to human), Xanthomonas maltophila, Aspergillus spp., and Cladosporium spp. (rarely pathogenic to human). Out of these, the dominant species in biofilm include Pseudomonas aeruginosa, Ochrobactrum anthropi, Alcaligenes denitrificans, Xanthomonas maltophila, and Vibrio harveyi.[[#References|[2]]] | Dominant bacteria are Staphylococcus spp.,[[#References|[5]]] [http://en.wikipedia.org/wiki/Pseudomonas_aeruginosa Pseudomonas aeruginosa], [[Ochrobactrum anthropi|Ochrobactrum anthropi]], [[Alcaligenes|Alcaligenes denitrificans]], [http://medical-dictionary.thefreedictionary.com/Xanthomonas+maltophilia Xanthomonas maltophila], and [http://en.wikipedia.org/wiki/Vibrio_harveyi Vibrio harveyi][[#References|[2]]], while dominant fungi are Aspergillus spp. And [http://en.wikipedia.org/wiki/Cladosporium Cladosporium spp.][[#References|[5]]] Opportunistic human pathogens included Staphylococcus spp., Pseudomonas aeruginosa, Ochrobactrum anthropi, Alcaligenes denitrificans (rarely pathogenic to human), Xanthomonas maltophila, Aspergillus spp., and Cladosporium spp. (rarely pathogenic to human). Out of these, the dominant species in biofilm include Pseudomonas aeruginosa, Ochrobactrum anthropi, Alcaligenes denitrificans, Xanthomonas maltophila, and Vibrio harveyi.[[#References|[2]]] | ||
===Fallouts (Dusts)=== | ===Fallouts (Dusts)=== | ||
Opportunistic pathogens identified are [http://en.wikipedia.org/wiki/Aspergillus_flavus Aspergillus flavus], and [http://en.wikipedia.org/wiki/Aspergillus_niger Aspergillus niger][[#References|[7]]]. Potential moderate toxin producers [http://en.wikipedia.org/wiki/Penicillium_chrysogenum Penicillium chrysogenum], and [http://www.moldbacteriaconsulting.com/tag/penicillium-brevicompactum Penicillium brevicompactum][[#References|[7]]] were also identified. | Opportunistic pathogens identified are [http://en.wikipedia.org/wiki/Aspergillus_flavus Aspergillus flavus], and [http://en.wikipedia.org/wiki/Aspergillus_niger Aspergillus niger][[#References|[7]]]. Potential moderate toxin producers [http://en.wikipedia.org/wiki/Penicillium_chrysogenum Penicillium chrysogenum], and [http://www.moldbacteriaconsulting.com/tag/penicillium-brevicompactum Penicillium brevicompactum][[#References|[7]]] were also identified. | ||
==Antimicrobial Techniques== | ==Antimicrobial Techniques== | ||
| Line 58: | Line 90: | ||
===Prevention of Antibiotic Resistance=== | ===Prevention of Antibiotic Resistance=== | ||
Ground-based researches on potential microbes assess the [http://en.wikipedia.org/wiki/Minimum_inhibitory_concentration minimum inhibitory concentration] (MIC) in microgravitaional conditions and develop proper [http://www.epa.gov/raf/microbial.htm microbial risk assessment] (MRA) that are used in determining efficient antibiotic usage and providing data base for future researches to minimize antibiotic resistance. [[#References|[12]]] | Ground-based researches on potential microbes assess the [http://en.wikipedia.org/wiki/Minimum_inhibitory_concentration minimum inhibitory concentration] (MIC) in microgravitaional conditions and develop proper [http://www.epa.gov/raf/microbial.htm microbial risk assessment] (MRA) that are used in determining efficient antibiotic usage and providing data base for future researches to minimize antibiotic resistance.[[#References|[12]]] | ||
==Current Research== | ==Current Research== | ||
| Line 66: | Line 99: | ||
===TianGong=== | ===TianGong=== | ||
The first Chinese space station, TianGong-1 (TG-1) was launched in September 2011.[[#References|[10]]] Two more space stations in this series, TG-2 and TG-3, are expected to be launched in subsequent years to researching on technologies required for long-term space missions.[[#References|[10]]] TianGong is projected to be upgraded to a large scale space station by 2020.[[#References|[10]]] | The first Chinese space station, TianGong-1 (TG-1) was launched in September 2011.[[#References|[10]]] Two more space stations in this series, TG-2 and TG-3, are expected to be launched in subsequent years to researching on technologies required for long-term space missions.[[#References|[10]]] TianGong is projected to be upgraded to a large scale space station by 2020.[[#References|[10]]] | ||
==References== | ==References== | ||
Latest revision as of 15:13, 1 October 2015
Introduction

Long-term space station missions to date include Salyut, Almaz, Skylab, Mir, ISS (International Space Station), and Tiangong, with the latter two still ongoing. Researches on the microbial communities which persist during these missions have demonstrated that space stations support a diverse microbial community. While most microbes are not harmful, routine microbial monitoring onboard space stations is still essential in preventing material biodeterioration[2] and maintaining crew health[3]. As currently identified, these microorganisms originate from Earth, with the main source being from crewmembers themselves and from materials manufactured on ground.[4] Environmental monitoring is important in identifying these microorganisms and providing data for future studies in attempt to minimize microbial threat to crewmembers’ performance and spacecrafts’ durability.
Physical Environment
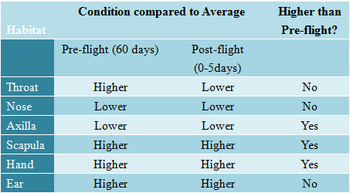
Microorganisms exist in the portable water, ventilating air, and on surfaces of space stations.[4] While these microorganisms share the same living space as the crewmembers, the crewmembers themselves also serve as incubators for various microbes.[5] Researchers have identified several strains of bacteria and fungi as opportunistic pathogen that may pose threats to crewmember. (refer to Key Microorganisms section) Moreover, they have also identified the type of microbes responsible for biodegradation of structural materials.[6] (refer to Biodegradation section)
Microbial Diversity and Interactions
Other than in portable water and air, bacteria and fungi typically co-exist in biofilms on surfaces, including water pipe lines.[7] Biofilms provides suitable growth conditions, and intrinsically contribute to antibiotic and biocide resistance[13] due to horizontal gene transfer of antibiotic resistance plasmid, and the fact that microbes positioned inside biofilms are harder to reach. One study shows that as much as 36 species of fungi and 58 species of bacteria can co-exist in a single biofilm.[16] However, the composition varies drastically due to dynamic (changing) environmental conditions, available nutrient (surface materials), and also the thickness of biofilm.[16] In addition, exchange of microflora carried by crewmembers travelling to and from the space craft also increases microbial diversity onboard space stations.[12]
Space Microbial Processes
Biodegradation
Biodegradation of the surfaces of spacecraft structural materials are results of colonization of bacteria and fungi.[2] While fungi are more abundant in early colonization and biofilm formation on polymeric materials, given sufficient humidity, subsequent microbes thrive in biofilms that forms at wide ranges of temperatures.[2] Biodegradation of bacteria and fungi predominantly occur on these surfaces due to the availability of carbon in surface finishings, as well as other nutrients present in structural materials[2] (polymeric-base and metal-based). In thin biofilms, oxic condition support aerobic biodegraders; in thicker biofilms, where oxygen diffusion is limited, anaerobic fermenters and metal reducers are come into play. Although these biodegradation of carbon (polymeric)-based and metal-based materials are due to similar hydrolysis reactions, the yielding products and degree of degradation are different depending on different microbes.[2]
Antibiotics Sensitivity
According to various in vitro studies, the microgravitaional environment in space enhances microbial growth.[1][12] Several factors were examined in a closed, isolated environment on ground-based labs, and the results show a decreased lag phase and increased final yield compared to normal-gravity conditions.[12] This enhanced microbial growth also contribute to increase of antibiotic resistance.[1][12] In addition to the confined, microgravitaional condition, various unknown factors onboard also provide distinctive selective pressures that favor certain microbes that eventually become harder to eliminate.[12]
Key Microorganisms
Microbe Identification Techniques (Onboard Environmental Monitering)
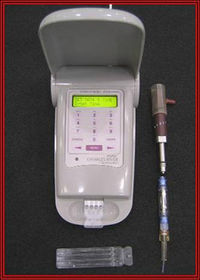
Most antimicrobial and environmental monitoring implementations on ISS are results of studies done by previous expedition, Mir and Skylab. Before December 2006, isolation of microorganisms depended on the growth of environmental isolates on agar media onboard space stations and the subsequent ground-based analysis back on Earth.[11] However, this method restricts the number identifiable microorganisms since more than 95% of environmental microbes do not grow on conventional agar growth media. Furthermore, accuracy is limited since months often passed before returning to Earth for further analysis.[11]
More recently, ISS launched Lab-On-A-Chip Application Development Portable Test System (LOCAD-PTS).[11] This new technology can be hand-held and allowed rapid microbial monitoring of bacteria and fungi since its implementation in December 2006.[11]
Portable Water
Dominant bacteria are Acidovorax spp., Afipia spp., Brevundimonas spp., Propionibacterium spp., Serratia spp.,[14] Sphingomonas spp., and Methylobacterium spp.,[5] and opportunistic human pathogens include Afipia spp., Brevundimonas spp., Propionibacterium spp., Serratia spp., and Sphingomonas spp. (rarely pathogenic to human).
Air
Dominant bacteria is Staphylococcus spp.[5], and dominant fungi are Aspergillus spp. and Penicillium spp..[5]
All are opportunistic human pathogens.
Surfaces
Dominant bacteria are Staphylococcus spp.,[5] Pseudomonas aeruginosa, Ochrobactrum anthropi, Alcaligenes denitrificans, Xanthomonas maltophila, and Vibrio harveyi[2], while dominant fungi are Aspergillus spp. And Cladosporium spp.[5] Opportunistic human pathogens included Staphylococcus spp., Pseudomonas aeruginosa, Ochrobactrum anthropi, Alcaligenes denitrificans (rarely pathogenic to human), Xanthomonas maltophila, Aspergillus spp., and Cladosporium spp. (rarely pathogenic to human). Out of these, the dominant species in biofilm include Pseudomonas aeruginosa, Ochrobactrum anthropi, Alcaligenes denitrificans, Xanthomonas maltophila, and Vibrio harveyi.[2]
Fallouts (Dusts)
Opportunistic pathogens identified are Aspergillus flavus, and Aspergillus niger[7]. Potential moderate toxin producers Penicillium chrysogenum, and Penicillium brevicompactum[7] were also identified.
Antimicrobial Techniques
The following antimicrobial methods focus on minimizing microbial colonization of the circulating air, portable water, and inside surfaces, as well as preventing the spread of antibiotic resistance. Data collected on ISS show that these methods were successful in reducing the microbial threat to crewmembers and the spacecraft.[9]
Ventilating Air
HEPA and onboard contamination controls effectively minimize contaminants and organic compounds in air.[9] This reduces the chance of microbial colonization by minimizing possible nutrient sources.
Portable Water
Iodide biocide is used to treat water sources on Earth before they were pumped onto space stations.[15]
Surfaces
The use of biocide, diiodomethyl-p-tolyl-sulfone, on surface finishings was proven effective in reducing microbial colonization.[13] However, current studies suggest that the best way to prevent microbial colonization on surfaces is to select for materials that inhibit biofilm formation and are less susceptible to potential microbial biodegraders.[2] Unfortunately, studies suggest that the material widely used in constructing spacecrafts, Fiber-reinforced polymeric composite materials (FRPCMs), is a potential site of microbe colonization due to its carbon content, is not resistant to biofilm formation, and is especially susceptible to fungal biodegradation.
Prevention of Antibiotic Resistance
Ground-based researches on potential microbes assess the minimum inhibitory concentration (MIC) in microgravitaional conditions and develop proper microbial risk assessment (MRA) that are used in determining efficient antibiotic usage and providing data base for future researches to minimize antibiotic resistance.[12]
Current Research
ISS
The core module of International Space Station was launched in 1998 with contributions from the United States, European Space Agency, Japan, Russia, and Canada.[8] One of the ongoing goals of the ISS expeditions is to research on methods of environmental microbial monitoring and controlling.[9]
TianGong
The first Chinese space station, TianGong-1 (TG-1) was launched in September 2011.[10] Two more space stations in this series, TG-2 and TG-3, are expected to be launched in subsequent years to researching on technologies required for long-term space missions.[10] TianGong is projected to be upgraded to a large scale space station by 2020.[10]
References
Text
[1] van Tongeren, S. P., Krooneman, J., Raangs, G. C., Welling, G. W. and Harmsen, H. J. M. “Microbial detection and monitoring in advanced life support systems like the International Space Station.” Microgravity Science and Technology, 2007, DOI: 10.1007/BF02911866
[2] Gu, J. “Microbial colonization of polymeric materials for space applications and mechanisms of biodeterioration: A review.” International Biodeterioration & Biodegradation, 2007, http://dx.doi.org/10.1016/j.ibiod.2006.08.010
[3] Castro, V. A., Thrasher, A. N., Healy, M., Ott C. M., and Pierson, D. L. “Microbial Characterization during the Early Habitation of the International Space Station.” Microbial Ecology, 2004, DOI: 10.1007/s00248-003-1030-y
[4] Pierson, D. L. “Microbial Contamination of Spacecraft.” Journal of the American Society for Gravitational and Space Biology, 2001, cited October 2012. http://gravitationalandspacebiology.org/index.php/journal/article/view/261
[5] Ilyin, V. K. “Microbiological status of cosmonauts during orbital spaceflights on Salyut and Mir orbital stations.” Acta Astronautica, 2005, http://dx.doi.org/10.1016/j.actaastro.2005.01.009
[6] Novikova, N., De Boever, P., Poddubko, S., Deshavaya, E., Polokarpov, N., Rakova, N., Coninx, I. and Mergeay, M. “Survey of environmental biocontamination on board the International Space Station.” Research in Microbiology, 2006, http://dx.doi.org/10.1016/j.resmic.2005.07.010
[7] Vesper, S. J., Wong, W., Kuo, C. M. and Pierson, D. L. “Mold species in dust from the International Space Station identified and quantified by mold-specific quantitative PCR.” Research in Microbiology, 2008, http://dx.doi.org/10.1016/j.resmic.2008.06.001
[8] NASA: International Space Station, cited October 2012. http://www.nasa.gov/mission_pages/station/main/index.html
[9] Robinson, J. A., Thumm, T. L. and Thomas, D. A. “NASA utilization of the International Space Station and the Vision for Space Exploration.” Acta Astronautica, 2007, http://dx.doi.org/10.1016/j.actaastro.2007.01.019
[10] Barbosa, R.C. “China launches TianGong-1 to mark next human space flight milestone.” NASASpaceFlight.com, 2011, cited October 2012. http://www.nasaspaceflight.com/2011/09/china-major-human-space-flight-milestone-tiangong-1s-launch/
[11] Gunter, D., Flores, G., Effinger, M., Maule, J., Wainwright, N., Steele, A., Damon, M., Wells, M., Williams, S., Morris, H. and Monaco, L. “Rapid Monitoring of Bacteria and Fungi aboard the International Space Station (ISS)” NASA Technical Reports Server (NTRS), 2009, http://naca.larc.nasa.gov/search.jsp?R=20090017684&qs=N%3D4294950110%2B4294848119%2B4294301177
[12] Klaus, D. M. and Howard, H. N. “Antibiotic efficacy and microbial virulence during space flight.” Trends in Biotechnology, 2006, http://dx.doi.org/10.1016/j.tibtech.2006.01.008
[13] Gu, J., Roman, M., Esselman, T. and Mitchell, R. “The role of microbial biofilms in deterioration of spacestation candidate materials.” International Biodeterioration & Biodegradation, 1998, http://dx.doi.org/10.1016/S0964-8305(98)80005-X
[14] La Duc, M. T., Sumner, R., Pierson, D., Venkat, P. and Venkateswaran, K. “Evidence of pathogenic microbes in the International Space Station drinking water: reason for concern?” UK Pubmed Central, 2004, cited on October 2012. http://ukpmc.ac.uk/abstract/MED/15880908/reload=0;jsessionid=40fVP5QP57u3BxVyDAfi.4
[15] La Duc, M. “103rd General Meeting of the American Society for Microbiology.” American Society for Microbiology, 2003, cited October 2012. http://www.asm.org/index.php?option=com_content&view=article&id=4205&title=How+Do+I+Get+My+First+Position+as+a+Microbiologist%3F+&Itemid=302
[16] Klintworth, R. and Reher, H. J. “Biological induced corrosion of materials II: New test methods and experiences from Mir station.” Acta Astronautica, 1999, http://dx.doi.org/10.1016/S0094-5765(99)00069-7
Image
[1] “STS-135 Shuttle Mission Imagery.” NASA, cited November 2012. http://spaceflight.nasa.gov/gallery/images/shuttle/sts-135/html/s135e011814.html
[2] “Astronaut Gloves Tested for Biological Contamination.” SPACE.com, cited November 2012. http://www.space.com/6486-astronaut-gloves-tested-biological-contamination.html