Purine alkaloid degradation: Difference between revisions
Meghandougan (talk | contribs) No edit summary |
No edit summary |
||
| Line 9: | Line 9: | ||
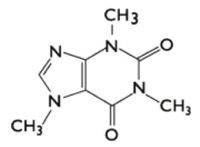
[http://en.wikipedia.org/wiki/Alkaloid Alkaloids] are secondary metabolites that have diverse structures (1), however, two commonalities underpin the chemical categorization: all alkaloids contain nitrogen and are classified as organic compounds. The purine alkaloids are a distinct form that have a nitrogen containing ring structure resembling that of purine. They are termed pseudoalkaloids, as they are not derived from amino acids like true alkaloids (2). The aromatic structure makes them recalcitrant to degradation. | [http://en.wikipedia.org/wiki/Alkaloid Alkaloids] are secondary metabolites that have diverse structures (1), however, two commonalities underpin the chemical categorization: all alkaloids contain nitrogen and are classified as organic compounds. The purine alkaloids are a distinct form that have a nitrogen containing ring structure resembling that of purine. They are termed pseudoalkaloids, as they are not derived from amino acids like true alkaloids (2). The aromatic structure makes them recalcitrant to degradation. | ||
[[File:Caffeinestructure.jpg]] | [[File:Caffeinestructure.jpg|200px|thumb|left|Figure 1: Structure of the purine alkaloid caffeine.]] | ||
| Line 18: | Line 18: | ||
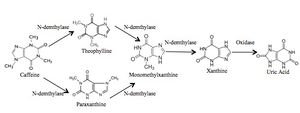
The two main enzymes involved in purine alkaloid catabolism are N-demethylase and xanthine oxidase. N-demethylase acts in the first steps of degradation, while xanthine oxidase catalyzes subsequent degradatory steps. Using caffeine (1,3,7-trimethylxanthine) as an example, purine alkaloid degradation is the process by which it is catabolized by N-1 and N-3 demethylases, releasing methyl groups to generate intermediates and eventually leading to xanthine production (4). This is followed by further degradation to the final product uric acid, catalyzed by xanthine oxidase. | The two main enzymes involved in purine alkaloid catabolism are N-demethylase and xanthine oxidase. N-demethylase acts in the first steps of degradation, while xanthine oxidase catalyzes subsequent degradatory steps. Using caffeine (1,3,7-trimethylxanthine) as an example, purine alkaloid degradation is the process by which it is catabolized by N-1 and N-3 demethylases, releasing methyl groups to generate intermediates and eventually leading to xanthine production (4). This is followed by further degradation to the final product uric acid, catalyzed by xanthine oxidase. | ||
[[File:Caffeine.jpg]] | [[File:Caffeine.jpg|thumb|300px|right|Figure 2: Caffeine degradation pathway by N-demethylases.]] | ||
| Line 30: | Line 30: | ||
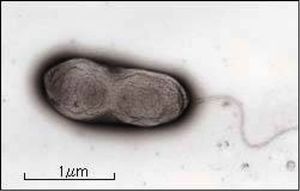
The process of purine alkaloid degradation has been found in multiple bacteria including ''[[Pseudomonas]]'', ''Serratia'', and ''[[Rhodococcus]]''. However the most well studied is ''P. putida''. It is a rod-shaped, flagellated, gram negative bacterium that possesses a diverse range of aerobic metabolism. For example, it has been observed living in caffeine concentrations of 50g/L (7). The strain ''Pseudomonas putida'' CBB5 was isolated from soil and found to catabolize caffeine by the mechanism described above using N-demethylases. The genes encoding enzymes required for caffeine degradation are related to a specific transmissible [http://en.wikipedia.org/wiki/Plasmid plasmid] found in caffeine-resistant strains of Pseudomonas. This plasmid may also be responsible for chemotaxis towards caffeine and related molecules (6). ''P. putida'' can also use the intermediates of caffeine degradation, theophylline and monomethylxanthine, for metabolism (10). This form of metabolism uses N-demethylation similar to that in caffeine metabolism with convergence of mechanisms at the xanthine stage. Because of its unique abilities to break down aromatic hydrocarbons and its nonpathogenic nature, this bacterium has high potential for [[bioremediation]]. | The process of purine alkaloid degradation has been found in multiple bacteria including ''[[Pseudomonas]]'', ''Serratia'', and ''[[Rhodococcus]]''. However the most well studied is ''P. putida''. It is a rod-shaped, flagellated, gram negative bacterium that possesses a diverse range of aerobic metabolism. For example, it has been observed living in caffeine concentrations of 50g/L (7). The strain ''Pseudomonas putida'' CBB5 was isolated from soil and found to catabolize caffeine by the mechanism described above using N-demethylases. The genes encoding enzymes required for caffeine degradation are related to a specific transmissible [http://en.wikipedia.org/wiki/Plasmid plasmid] found in caffeine-resistant strains of Pseudomonas. This plasmid may also be responsible for chemotaxis towards caffeine and related molecules (6). ''P. putida'' can also use the intermediates of caffeine degradation, theophylline and monomethylxanthine, for metabolism (10). This form of metabolism uses N-demethylation similar to that in caffeine metabolism with convergence of mechanisms at the xanthine stage. Because of its unique abilities to break down aromatic hydrocarbons and its nonpathogenic nature, this bacterium has high potential for [[bioremediation]]. | ||
[[File:Pputida.jpg]] | [[File:Pputida.jpg|thumb|300px|right|Figure 3: ''Pseudomonas Putida''.]] | ||
Latest revision as of 04:07, 27 December 2012
Introduction
Purine alkaloids are produced by plants, examples of which include caffeine, cocaine and nicotine. Degradation of purine alkaloids occurs in plants, fungi and bacteria. In bacteria, it is the process by which caffeine (1,3,7-trimethylxanthine) and other purine alkaloids are catabolized by N-demethylases, producing xanthine and then further degraded by oxidases. An alternative pathway involves solely oxidative methods. Caffeine in particular can be quite toxic to some microbes; this has been shown in Escherichia coli DH5α (3). At the other end of the spectrum, there are some microbes such as Pseudomonas Putida that use caffeine for metabolism as their main carbon and nitrogen source. These organisms are often closely associated with plants and purine alkaloid synthesizing organisms. Microorganisms such as P. putida are therefore of interest for industrial processes like the decaffeination of beverages and coffee pulp pollution (5).
Purine Alkaloid Structure
Alkaloids are secondary metabolites that have diverse structures (1), however, two commonalities underpin the chemical categorization: all alkaloids contain nitrogen and are classified as organic compounds. The purine alkaloids are a distinct form that have a nitrogen containing ring structure resembling that of purine. They are termed pseudoalkaloids, as they are not derived from amino acids like true alkaloids (2). The aromatic structure makes them recalcitrant to degradation.
Mechanism of Purine Alkaloid Degradation
The two main enzymes involved in purine alkaloid catabolism are N-demethylase and xanthine oxidase. N-demethylase acts in the first steps of degradation, while xanthine oxidase catalyzes subsequent degradatory steps. Using caffeine (1,3,7-trimethylxanthine) as an example, purine alkaloid degradation is the process by which it is catabolized by N-1 and N-3 demethylases, releasing methyl groups to generate intermediates and eventually leading to xanthine production (4). This is followed by further degradation to the final product uric acid, catalyzed by xanthine oxidase.
An alternative pathway involves solely oxidative methods. It involves a different oxidase enzyme, caffeine oxidase, which functions in a completely separate process where caffeine is catabolized directly to 1,3,7-trimethyluric acid instead of passing through a pathway of intermediates (4). This course of degradation does not involve N-demethylases and is therefore entirely oxidative. These mechanisms of catabolizing the purine alkaloid caffeine are present in plants, fungi and bacterial microorganisms.
Key Microorganisms
The process of purine alkaloid degradation has been found in multiple bacteria including Pseudomonas, Serratia, and Rhodococcus. However the most well studied is P. putida. It is a rod-shaped, flagellated, gram negative bacterium that possesses a diverse range of aerobic metabolism. For example, it has been observed living in caffeine concentrations of 50g/L (7). The strain Pseudomonas putida CBB5 was isolated from soil and found to catabolize caffeine by the mechanism described above using N-demethylases. The genes encoding enzymes required for caffeine degradation are related to a specific transmissible plasmid found in caffeine-resistant strains of Pseudomonas. This plasmid may also be responsible for chemotaxis towards caffeine and related molecules (6). P. putida can also use the intermediates of caffeine degradation, theophylline and monomethylxanthine, for metabolism (10). This form of metabolism uses N-demethylation similar to that in caffeine metabolism with convergence of mechanisms at the xanthine stage. Because of its unique abilities to break down aromatic hydrocarbons and its nonpathogenic nature, this bacterium has high potential for bioremediation.
Significance
One of the characteristics of alkaloids is that they act on the central nervous system of humans and other mammals (2). Activity of alkaloids can affect major chemical transmitters such as epinephrine. This is certainly true of caffeine, however, this can cause antagonist effects such as cardiovascular problems and pregnancy complications (4). Because of this, it is beneficial to decaffeinate products. Conventional techniques of decaffeination can be expensive, toxic and cause products to be less tasteful (6). Therefore, the use of microbial decaffeination methods can be advantageous to avoid these problems. Another usage for biological decaffeination by microbes is the removal of caffeine from coffee and tea pulps. These are generated during the production of coffee and tea and discharged into water sources where they can be used as nutritional resources, but only if they are void of caffeine. In this sense, decaffeination techniques are an environmental advantage (5).
Current Research
Defining the enzymology behind purine alkaloid degradation in microbes is essential in being able to implement biodecaffeination processes. Previously, metabolism of caffeine was credited to one broadly specific N-demethylase, but recent studies have shown that it is actually accomplished by two independent enzymes. These enzymes are actually suggested to be monooxygenases with N-1 and N-3 demethylase activity. The genes for these NdmA and NdmB enzymes have been pinpointed to a specific genomic DNA fragment, which also contains genes for NdmD (the enzyme responsible for electron transfer from NADH) and NdmC (an N-7 demethylase) (8). The knowledge of these enzymes could have vast bioremediation applications, as well as provide a leeway to the discovery of new N-demethylases.
References
(1) Anaya, A., Cruz-Ortega, R., Waller, G., “Metabolism and Ecology of Purine Alkaloids.” Frontiers in Bioscience, 2006.
(2) Aniszewski, T., “Alkaloids – Secrets of Life.” Elsevier, 2007.
(3) Dash, S., Gummadi, S., “Inhibitory Effect of Caffeine on Growth of Various Bacterial Strains.” Research Journal of Microbiology, 2008.
(4) Dash, S., Gummadi, S., “Catabolic pathways and biotechnological applications of microbial caffeine degradation.” Biotechnology Letters, 2006.
(5) Gokulakrishnan, S., Chandraraj, K., Gummadi, S., “Microbial and enzymatic methods for the removal of caffeine.” Enzyme and Microbial Technology, 2005.
(6) Gummadi, S., Bhavya, B., Ashok, N., “Physiology, Biochemistry and possible applications of microbial caffeine degradation.” Applied Microbiology Biotechnology, 2011.
(7) Mazzafera, P., “Degradation of caffeine by microorganisms and potential use of decaffeinated coffee husk and pulp in animal feeding.” Scientia Agricola, 2002.
(8) Summers, R., Louie, T., Yu, C., Gakhar, L., Louie, K., Subramanian, M., “Novel, Highly Specific N-Demethylases Enable Bacteria To Live on Caffeine and Related Purine Alkaloids.” Journal of Bacteriology, 2012.
(9) Summers, R., Louie, T., Yu, C., Subramanian, M., “Characterization of a broad-specificity non-haem iron N-demethylase from Pseudomonas putida CBB5 capable of utilizing several purine alkaloids as sole carbon and nitrogen source.” Microbiology, 2010.
(10) Yu, C., Louie, T., Summers, R., Kale, Y., Gopishetty, S., Subramanian, M., “Two Distinct Pathways for Metabolism of Theophylline and Caffeine are Coexpressed in Pseudomonas putida CBB5.” Journal of Bacteriology, 2009.