HIV Envelope and Cell Fusion: Difference between revisions
| Line 20: | Line 20: | ||
<br>Introduce the topic of your paper. What microorganisms are of interest? Habitat? Applications for medicine and/or environment?<br> | <br>Introduce the topic of your paper. What microorganisms are of interest? Habitat? Applications for medicine and/or environment?<br> | ||
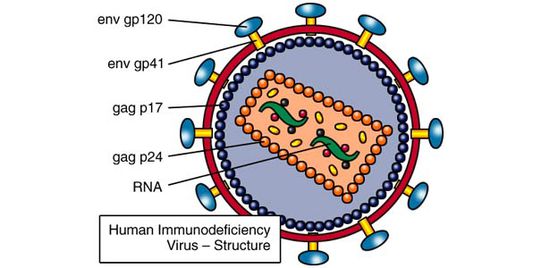
[[File:HIV_Structure_Image.jpg|thumb||550px||right|Figure X. Diagram of an HIV virion, with major protein components labeled. Image credit: http://www.avert.org/hiv-structure-and-life-cycle.htm.]] | [[File:HIV_Structure_Image.jpg|thumb||550px||right|Figure X. Diagram of an HIV virion, with major envelope and capsid protein components labeled. Image credit: http://www.avert.org/hiv-structure-and-life-cycle.htm.]] | ||
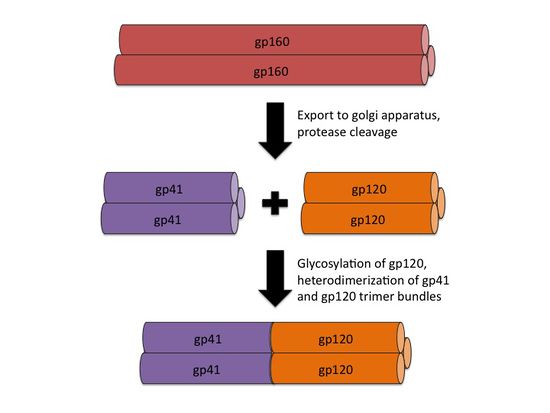
[[File:HIV_gp160_processing.jpg|thumb||550px||right|Figure X. An illustration showing the processing of the gp160 intermediate into gp41 and gp120 homotrimer bundles, followed by heterodimerization. Image credit: Ian Perrone.]] | [[File:HIV_gp160_processing.jpg|thumb||550px||right|Figure X. An illustration showing the processing of the gp160 intermediate into gp41 and gp120 homotrimer bundles, followed by heterodimerization. Image credit: Ian Perrone.]] | ||
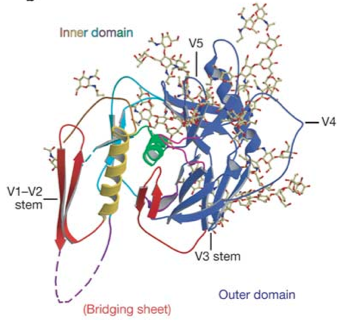
[[File:gp120_structure_image.png|thumb||550px||right|Figure X. Crystal structure of SIV gp120. While not exactly the same in sequence as HIV gp120, SIV gp120 includes the same conserved structures. Image credit: Chen et al., 2005.]] | [[File:gp120_structure_image.png|thumb||550px||right|Figure X. Crystal structure of SIV gp120. While not exactly the same in sequence as HIV gp120, SIV gp120 includes the same conserved structures. Image credit: Chen et al., 2005. (http://www.nature.com/nature/journal/v433/n7028/abs/nature03327.html)]] | ||
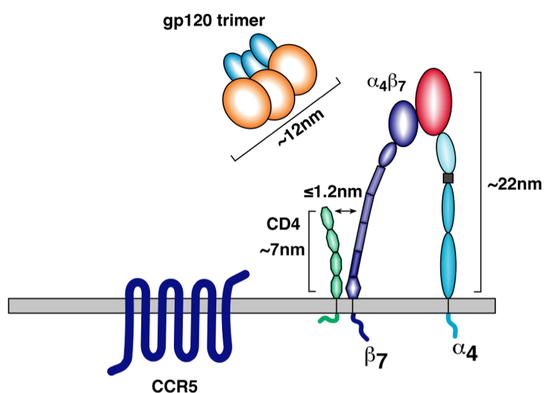
[[File:a4b7_diagram.png|thumb||550px||right|Figure X. Diagram showing the spatial relationship of α4β7 integrin and the main HIV coreceptors, CD4 and CCR5, on the surface of a α4β7 -expressing T-cell. Image credit: Cicala et al., 2010.]] | [[File:a4b7_diagram.png|thumb||550px||right|Figure X. Diagram showing the spatial relationship of α4β7 integrin and the main HIV coreceptors, CD4 and CCR5, on the surface of a α4β7 -expressing T-cell. Image credit: Cicala et al., 2010. (http://www.biomedcentral.com/content/pdf/1479-5876-9-S1-S2.pdf)]] | ||
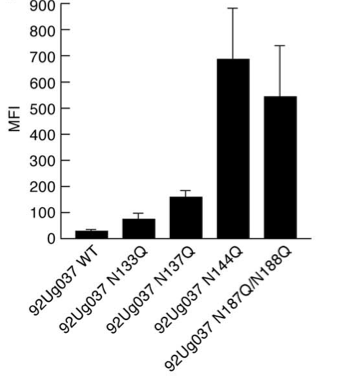
[[File:gp120_PNG_mutations.png|thumb||550px||right|Figure X. Graph showing mean fluorescence intensity (MFI), an indicator of α4β7 binding, as a function of PNG deletions from gp120. PNG mutations were achieved by replacing the indicated asparagine residues of the V1 and V2 loops with glutamine. Image credit: Nawaz et al., 2011.]] | [[File:gp120_PNG_mutations.png|thumb||550px||right|Figure X. Graph showing mean fluorescence intensity (MFI), an indicator of α4β7 binding, as a function of PNG deletions from gp120. PNG mutations were achieved by replacing the indicated asparagine residues of the V1 and V2 loops with glutamine. Image credit: Nawaz et al., 2011. (http://www.plospathogens.org/article/info%3Adoi%2F10.1371%2Fjournal.ppat.1001301#ppat-1001301-g007)]] | ||
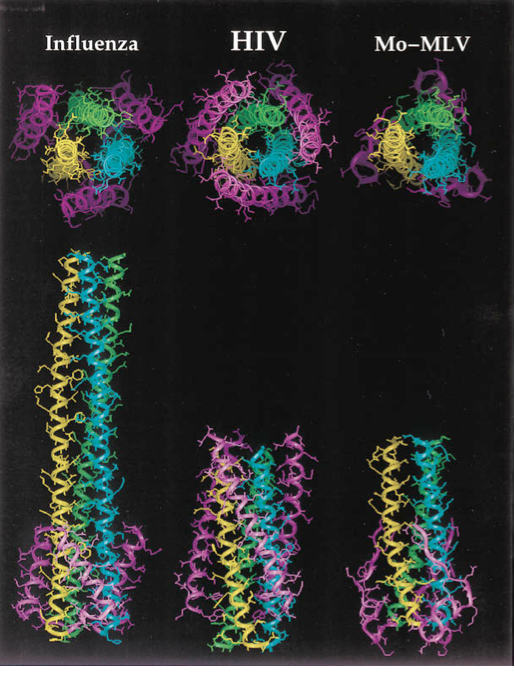
[[File:gp41_comparison.png|thumb||550px||right|Figure X. Diagram showing a structural comparison of the gp41 ectodomain with functionally similar regions of the envelope fusion proteins of influenza and murine leukemia virus, Mo-MLV. The high degree of conservation of the coiled coil motif is a strong indicator of its effectiveness as a fusion initiation mechanism. Image credit: Chan et al., 1997.]] | [[File:gp41_comparison.png|thumb||550px||right|Figure X. Diagram showing a structural comparison of the gp41 ectodomain with functionally similar regions of the envelope fusion proteins of influenza and murine leukemia virus, Mo-MLV. The high degree of conservation of the coiled coil motif is a strong indicator of its effectiveness as a fusion initiation mechanism. Image credit: Chan et al., 1997. (http://www.sciencedirect.com/science/article/pii/S0092867400802056)]] | ||
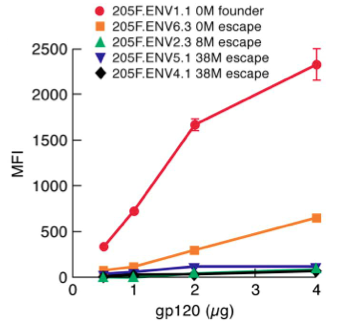
[[File:gp120_escape.png|thumb||550px||right|Figure X. Graph showing the α4β7 binding affinity (measured in terms of MFI) of HIV clinical isolates collected from a female patient over the course of 38 months. Image credit: Nawaz et al., 2011.]] | [[File:gp120_escape.png|thumb||550px||right|Figure X. Graph showing the α4β7 binding affinity (measured in terms of MFI) of HIV clinical isolates collected from a female patient over the course of 38 months. Image credit: Nawaz et al., 2011. (http://www.plospathogens.org/article/info%3Adoi%2F10.1371%2Fjournal.ppat.1001301#ppat-1001301-g007)]] | ||
[[File:|thumb||550px||right|Figure X. ]] | [[File:|thumb||550px||right|Figure X. ]] | ||
Revision as of 04:00, 22 April 2014
Introduction
By Ian Perrone
At right is a sample image insertion. It works for any image uploaded anywhere to MicrobeWiki. The insertion code consists of:
Double brackets: [[
Filename: PHIL_1181_lores.jpg
Thumbnail status: |thumb|
Pixel size: |300px|
Placement on page: |right|
Legend/credit: Electron micrograph of the Ebola Zaire virus. This was the first photo ever taken of the virus, on 10/13/1976. By Dr. F.A. Murphy, now at U.C. Davis, then at the CDC.
Closed double brackets: ]]
Other examples:
Bold
Italic
Subscript: H2O
Superscript: Fe3+
Introduce the topic of your paper. What microorganisms are of interest? Habitat? Applications for medicine and/or environment?
[[File:|thumb||550px||right|Figure X. ]]
[[File:|thumb||550px||right|Figure X. ]]
[[File:|thumb||550px||right|Figure X. ]]
Section 1
Include some current research, with at least one figure showing data.