Rhodococcus ruber: Difference between revisions
| Line 28: | Line 28: | ||
[[ File:DSM_44190.jpg | thumb| 200px| right| Figure 2: ''Rhodococcus ruber'' cultured on a trypticase soy broth agar [https://bacdive.dsmz.de/strain/10942] ]] | [[ File:DSM_44190.jpg | thumb| 200px| right| Figure 2: ''Rhodococcus ruber'' cultured on a trypticase soy broth agar [https://bacdive.dsmz.de/strain/10942] ]] | ||
Historically, the genus ''Rhodococcus'' was first defined by Zopf in 1891. ''Nocardia rubra'', or what is known today as ''Rhodococcus ruber'', was isolated from a soil sludge by Kruse in 1896. However, there were issues with classification due to morphological and staining identification techniques. In 1977, Goodfellow and Alderson did a complete reclassification of the genus ''Rhodococcus'', which resulted in ''Norcordia rubra'' being amended to ''Rhodococcus ruber''. | Historically, the genus ''Rhodococcus'' was first defined by William Zopf in 1891. ''Nocardia rubra'', or what is known today as ''Rhodococcus ruber'', was isolated from a soil sludge by William Kruse in 1896. However, there were issues with classification due to morphological and staining identification techniques. In 1977, Micheal Goodfellow and Grace Alderson did a complete reclassification of the genus ''Rhodococcus'', which resulted in ''Norcordia rubra'' being amended to ''Rhodococcus ruber''. | ||
The term ''Rhodococcus'' is from the Greek words “rhodon” and “coccus” meaning “rose” and “grain“ respectively. ''R. ruber's'' morphological structure matches the origins of its name: rods and cocci. ''R. ruber'' first starts as long rods, then breaks off into short rods and cocci throughout different growth phases. ''R. ruber'' is characteristically a gram positive, non-motile, non-spore forming bacteria. It has diverse metabolic and nutritional capabilities depending on the strain, such as utilizing gaseous hydrocarbons, aromatic hydrocarbons, complex polymers, and steroids for carbon and energy sources. | The term ''Rhodococcus'' is from the Greek words “rhodon” and “coccus” meaning “rose” and “grain“ respectively. ''R. ruber's'' morphological structure matches the origins of its name: rods and cocci. ''R. ruber'' first starts as long rods, then breaks off into short rods and cocci throughout different growth phases. ''R. ruber'' is characteristically a gram positive, non-motile, non-spore forming bacteria. It has diverse metabolic and nutritional capabilities depending on the strain, such as utilizing gaseous hydrocarbons, aromatic hydrocarbons, complex polymers, and steroids for carbon and energy sources. | ||
Revision as of 22:32, 29 April 2020
Classification
Domain: Bacteria
Phylum: Actinobacteria
Class: Actinobacteria
Order: Corynbacteriales
Family: Nocardiaceae
Genus: Rhodococcus
Species
|
NCBI: [2] |
Rhodococcus ruber
Description and Significance
Historically, the genus Rhodococcus was first defined by William Zopf in 1891. Nocardia rubra, or what is known today as Rhodococcus ruber, was isolated from a soil sludge by William Kruse in 1896. However, there were issues with classification due to morphological and staining identification techniques. In 1977, Micheal Goodfellow and Grace Alderson did a complete reclassification of the genus Rhodococcus, which resulted in Norcordia rubra being amended to Rhodococcus ruber.
The term Rhodococcus is from the Greek words “rhodon” and “coccus” meaning “rose” and “grain“ respectively. R. ruber's morphological structure matches the origins of its name: rods and cocci. R. ruber first starts as long rods, then breaks off into short rods and cocci throughout different growth phases. R. ruber is characteristically a gram positive, non-motile, non-spore forming bacteria. It has diverse metabolic and nutritional capabilities depending on the strain, such as utilizing gaseous hydrocarbons, aromatic hydrocarbons, complex polymers, and steroids for carbon and energy sources.
R. ruber is unique in that it is ubiquitous in many different habitats whether that is pristine, contaminated, temperature, extreme. It is able to persist and tolerate stressful conditions, such as limited oxygen and nutrients. Given its metabolic and habitat versatility, R. ruber is a good candidate for bioremediation, biocatalysts, and other biotechnological uses to naturally degrade pollutants or remedy contaminated ecosystems.
Genome Structure
12 genome studies have been reported for the following strains of Rhodococcus ruber: R. ruber str. YC-YT1, R. ruber str. YYL, R. ruber str. P14, R. ruber str. SD3, R. ruber str. R1, R. ruber str. DSM 43338, R. ruber str. P25, R. tuber IEGM 231, R. ruber str. OA1, R. ruber str. BKS 20-38, R. ruber str. Chol-4, and R. ruber str. NCRX 15591.
The genome of Rhodococcus ruber consists of one circular chromosome with an average size of 5.57 Mb. The genome size varies for every strain and larger genome sizes can be attributed to the presence of circular plasmids. The strains of R. ruber that contain plasmids are YC-TC1, YYL, and R1. The average amount of protein coding genes is 4932 and the average G+C content is 70.49%. A high guanine-cytosine content is characteristic of Rhodococcus species to provide DNA stability in varying conditions.
The genome sequences of R. ruber are helpful in identifying the genes necessary for metabolic pathways and cycles that R. ruber employs including the tricarboxylic acid cycle, pentose phosphate pathway, glycolysis, and gluconeogenesis. Additionally, the beta oxidation pathway and alkane degradation pathway are utilized to break down more complex xenobiotic compounds before the classic metabolic pathways are used. The genome highlights multiple metabolic gene clusters that are used for degradation of aromatic compounds, steroids, hydrocarbons, and xenobiotic polymers that R. ruber uses for carbon and energy sources. Some strains of R. ruber contain redundant genes which contributes to their versatile metabolic abilities with various nutrient sources.
Cell Structure and Metabolism
It is a gram positive, non-motile, non-spore forming.
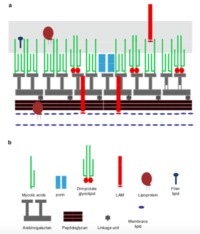
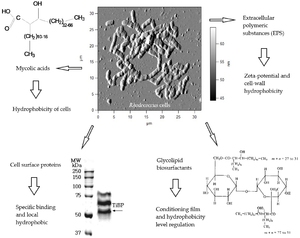
Mycolic acids= Long chain fatty acids
Mycolic acids with chain lengths of C42-C48; much longer chains than other species of Rhodococcus
Cell envelope lipids
Have pronounced surfactant properties that facilitate the growth of the bacteria on hydrophobic substrates
Laccases are phenol oxidases, which are part of a group of multi-copper enzymes They function by promoting biotic oxidation Adding copper on PE cultures, which enhanced degradation by 75% compared to non-amended cultures Copper helps induce laccase’s oxidizing activity
Ecological Impacts
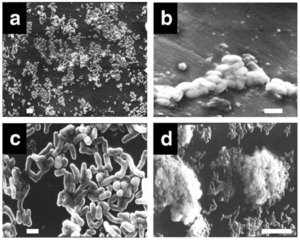
R. ruber will form a biofilm
This has a higher metabolic activity than if it was suspended and the solid surface serves as a substrate for R. ruber, so carbon availability is greater in a biofilm
The colonization of R. ruber to PE is nonspecific
Adheres to solid surface of PE, becomes differentiated from planktonic state, initiates formation of 3D structures of microcolonies, then further organized into multicellular mushroom-like structures that last for 60 + days
Carbon starvation enhanced the hydrophobic interactions, biofilm development, which improved biodegradation of PE
References
[1]| Alvarez, H. M. Biology of Rhodococcus; Springer, 2010; Vol. 16.
[11] Rhodococcus ruber (ID 11562) https://www.ncbi.nlm.nih.gov/genome/?term=txid1830[Organism:noexp] (accessed 2020).
Author
Page authored by Hannah von Werder, student of Prof. Jay Lennon at Indiana University.