Salmonella enterica NEU2011: Difference between revisions
No edit summary |
|||
| Line 35: | Line 35: | ||
===Metabolism=== | ===Metabolism=== | ||
This bacteria exhibits mixed-acid heterofermentation of glucose to produce ATP. This produces CO2 and H2 with various acid end-products, such as formate, acetate, lactate, and succinate [http://www.ncbi.nlm.nih.gov/books/NBK7919/#A398 (12).] <i>S. enteric</i> also has the ability to to extract iron from host proteins by siderophores, which are made by the cell when iron concentration is low. There is also evidence that suggests siderophores contribute to virulence [http://www.ncbi.nlm.nih.gov/books/NBK8526/#A575 (13).] | This bacteria exhibits mixed-acid heterofermentation of glucose to produce ATP. This produces CO2 and H2 with various acid end-products, such as formate, acetate, lactate, and succinate [http://www.ncbi.nlm.nih.gov/books/NBK7919/#A398 (12).] <i>S. enteric</i> also has the ability to to extract iron from host proteins by siderophores, which are made by the cell when iron concentration is low. There is also evidence that suggests siderophores contribute to virulence [http://www.ncbi.nlm.nih.gov/books/NBK8526/#A575 (13).] | ||
<i>Salmonela enterica</i> can also feed on maltose and maltodextrins through a type I ATP-binding cassette(ABC) transporter. This system is encoded on the maltose regulon. Maltose is imported into the cell by crossing the outer membrane through a homotrimeric maltoporin, the maltose then binds to maltose-binding protein, which along with a protein with pore-forming subunits and an ATP-binding subunit is tranlocated across the inner-membrane where is can be used by the cell [http://www.ncbi.nlm.nih.gov/pubmed/20659291 (14).] | <i>Salmonela enterica</i> can also feed on maltose and maltodextrins through a type I, ATP-binding cassette(ABC) transporter. This system is encoded on the maltose regulon. Maltose is imported into the cell by crossing the outer membrane through a homotrimeric maltoporin, the maltose then binds to maltose-binding protein, which along with a protein with pore-forming subunits and an ATP-binding subunit is tranlocated across the inner-membrane where is can be used by the cell [http://www.ncbi.nlm.nih.gov/pubmed/20659291 (14).] | ||
==Ecology== | ==Ecology== | ||
Revision as of 20:52, 1 April 2011
Classification
Higher Order Taxa
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae
Genus & Species
Salmonella enterica
Description and Significance
Salmonella enterica is a gram-negative, rod-shaped, flagellated bacterium that is of interest due to its ability to cause infectious disease in humans and animals. Salmonella is facultative, being capable of both aerobic respiration for the production ATP as well as fermentation in the absence of oxygen. Dr. Daniel E. Salmon was the first person to identify Salmonella species over 100 years ago, currently there are approximately 2,300 serotypes known. Salmonella infection is known as Salmonellosis; Salmonella enterica subspecies enterica serovar thyphi is a subspecies that is responsible for typhoid fever in humans by invading the gastrointestinal tract. Much of the current understanding of cellular and molecular functions in a cell has been made possible by using Salmonella. For example, Salmonella enterica subspecies enterica serovar Typhimurium, was the first bacteria used to observe transduction by Zinder and Lederberg in 1952. Since then a vast number of discoveries have been made using Salmonella ranging from gene regulation mechanisms, to cell surface antigen interactions with immune cells (1),(2), (3).
Genome Structure
The genome of Salmonella enterica serovar Typhi Ty2 is almost 4.8 megabases long and is comprised of 52.05% G-C base pairs. There are 4323 genes, all of which encode for functioning proteins. There are 78 tRNA genes contained within the genome along with 22 rRNA genes. The genome of S. enterica differs from that of Escherichia coli by ~10% and all serovar subtypes have as much as a 1% difference amongst each other in regards to net base pair differences. (4)
The genome can include plasmids but all subtypes have one main circular chromosome. (5)
The genome contains some outlying genes which are mostly unrelated to the genes contained within the base genome; some of these genes contain increased G-C base pairs which suggests that they were acquired more recently than most of the genome through a horizontal gene transfer. An example of these potentially newer genes would be the Salmonella pathogenicity island genes (abbreviated to "SPI", the two primary SPIs are known as SPI-1 and SPI-2) which affect the survival of S. enterica in eukaryotic cells. The study of S. Typhi and its single nucleotide polymorphisms (SNPs) has shown that it may be as young as 30,000 years old which would put its divergence far downfield from the divergence between Salmonella enterica and Escherichia coli. (5)
Cell Structure and Metabolism
S. enteric in a gram-negative, peritrichously flagellated, faculatively anaerobic, bacilli.
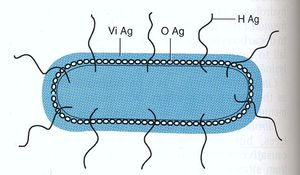
Antigens
Salmonella species are characterized by three antigens present:
- H antigen: This is the flagellar antigen. It occurs in either phase 1 or phase 2 and can alternate which is expressed (6),(7).
- O antigen: This is the somatic antigen. It is present on the outer membrane and its specificity is determined by the nature of the repeating units in the outer O-polysaccharide chain (6).
- Vi antigen: This is the virulence antigen. It is a capsular polysaccharide overlays the O antigen. It is not possessed by all serovars. This capsule is not necessary for infection, increase the infectivity by making it less detectable by the body’s immune system (8).
Fimbriae
S. enterica also possesses type I fimbriae, typical of other enteric bacteria, which aid in binding to epithelium cells (9). They are shorter than flagella but have a similar peritrichous distribution. They are made up of major and minor protein subunits that form a cylinder around a hollow core (10). Not all strains of S. enterica express fimbriae at the same time, but the fimbriaeted ones appear to be more virulent (11).
Metabolism
This bacteria exhibits mixed-acid heterofermentation of glucose to produce ATP. This produces CO2 and H2 with various acid end-products, such as formate, acetate, lactate, and succinate (12). S. enteric also has the ability to to extract iron from host proteins by siderophores, which are made by the cell when iron concentration is low. There is also evidence that suggests siderophores contribute to virulence (13). Salmonela enterica can also feed on maltose and maltodextrins through a type I, ATP-binding cassette(ABC) transporter. This system is encoded on the maltose regulon. Maltose is imported into the cell by crossing the outer membrane through a homotrimeric maltoporin, the maltose then binds to maltose-binding protein, which along with a protein with pore-forming subunits and an ATP-binding subunit is tranlocated across the inner-membrane where is can be used by the cell (14).
Ecology
The main habitat where Salmonella is found is in the intestines of animals and humans. It is also likely to find them inhabiting soil, water, and some plants due to human or animal excretion into the environment (1). Salmonella enterica may be found in many foods due to fecal contamination by infected or carrier animals or humans (14). Most commonly, food that is grown in soil that is treated with manure to increase the fertility of that land can be contaminated with S. enteric as well as other pathogenic microbes (15).
Pathology
Infectious diseases caused by Salmonella enterica depend largely on secreted proteins and adhesions from both fimbrial and non-fimbrial sources to create a biofilm and contact with the host cell (16). Bacterial cells enter the cells that line the intestine, and the host cell’s membrane ruffles as a response to the initial attachment of the bacteria. Ruffling is related to a triggering reaction that is results in macropinocytosis. The vesicles are fairly large during macropinocytosis, providing an efficient route for non-selective endocytosis of solute macromolecules (17). The entrance of the bacterial cells briefly harms the microvilli on the cell surface. Following this disruption, white blood cells flood the mucosa which alters the homeostasis between absorption and secretion in the body (18).
Bacterial cells harm the host cells by causing intracellular unrestricted calcium to rise and dislocate the cytoplasm. The infectious cells can then be transferred to the liver or spleen, where it multiplies (6). S.enterica either return to the host’s intestinal tract, or can be defecated. In addition to food contamination, further infection often occurs from infested feces by polluted water, soil, or other unclean environments.
Salmonella enterica is the cause of many cases of abdominal complications like gastroenteritis. When the balance between absorption and secretion is altered, initial symptoms include abdominal cramps, diarrhea, fever, and nausea. Most cases caused by food contamination resolve themselves within one week’s time, but more serious cases can often need fluoroquinolone or cephalosporins, both are broad-spectrum antibiotics that can respectively kill the organism or stop the cell wall from forming (19).
Current Research
Cool Factor
References
9. Madigan M, Martinko J, Dunlap P, Clark D. Brock Biology of Microorganisms. San Francisco (CA): Pearson Education; 2009.
11. Mahon CR, Manuselis G. Enterobacteriaceae. In: Mahon CR, Manuselis G, editors. Textbook of Diagnostic Microbiology. Philadelphia: W.B. Saunders; 1995.
Edited by Abigail Bootes, Matthew Brennan, Alex Birch, and Danielle Laramee for BIOL2321 Microbiology, 2011, Northeastern University